Difference between revisions of "YBR284W"
SGDwikiBot (talk | contribs) (Automated import of articles) |
(→UW-Stout/Copper FA21) |
||
| (26 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
{|{{Prettytable}} align = 'right' width = '200px' | {|{{Prettytable}} align = 'right' width = '200px' | ||
|- | |- | ||
| − | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http:// | + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http://www.yeastgenome.org/cgi-bin/locus.pl?dbid=S000000488 YBR284W] |
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | ||
| − | |nowrap| Chr II: | + | |nowrap| Chr II:771239..773632 |
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Primary SGDID''' || S000000488 | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Primary SGDID''' || S000000488 | ||
|} | |} | ||
<br> | <br> | ||
| − | '''Description of YBR284W:''' | + | '''Description of YBR284W:''' Protein of unknown function; some similarity to AMP deaminases but lacks key catalytic residues and does not rescue purine nucleotide metabolic defect of quadruple aah1 ade8 amd1 his1 mutant; null mutant exhibits longer telomeres, altered Ty mobility, decreased resistance to rapamycin and wortmannin, and synthetic phenotype with expression of alpha-synuclein; induced in response to hydrostatic pressure; not an essential gene<ref name='S000114973'>Gatbonton T, et al. (2006) Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet 2(3):e35 {{SGDpaper|S000114973}} PMID 16552446</ref><ref name='S000045570'>Holmstrom K, et al. (1994) The sequence of a 32,420 bp segment located on the right arm of chromosome II from Saccharomyces cerevisiae. Yeast 10 Suppl A:S47-62 {{SGDpaper|S000045570}} PMID 8091861</ref><ref name='S000124801'>Iwahashi H, et al. (2003) Piezophysiology of genome wide gene expression levels in the yeast Saccharomyces cerevisiae. Extremophiles 7(4):291-8 {{SGDpaper|S000124801}} PMID 12910389</ref><ref name='S000122748'>Maxwell PH and Curcio MJ (2007) Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for regulation of mammalian retroviruses. Eukaryot Cell 6(7):1069-80 {{SGDpaper|S000122748}} PMID 17496126</ref><ref name='S000131087'>Saint-Marc C, et al. (2009) Phenotypic consequences of purine nucleotide imbalance in Saccharomyces cerevisiae. Genetics 183(2):529-38, 1SI-7SI {{SGDpaper|S000131087}} PMID 19635936</ref><ref name='S000113969'>Willingham S, et al. (2003) Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science 302(5651):1769-72 {{SGDpaper|S000113969}} PMID 14657499</ref><ref name='S000081669'>Xie MW, et al. (2005) Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method. Proc Natl Acad Sci U S A 102(20):7215-20 |
{{SGDpaper|S000081669}} PMID 15883373</ref> | {{SGDpaper|S000081669}} PMID 15883373</ref> | ||
<br> | <br> | ||
| Line 29: | Line 29: | ||
==Community Commentary== | ==Community Commentary== | ||
{{CommentaryHelp}} | {{CommentaryHelp}} | ||
| + | |||
| + | {{UW-Stout}} | ||
| + | ===[[UW-Stout/Heat Shock FA21]]=== | ||
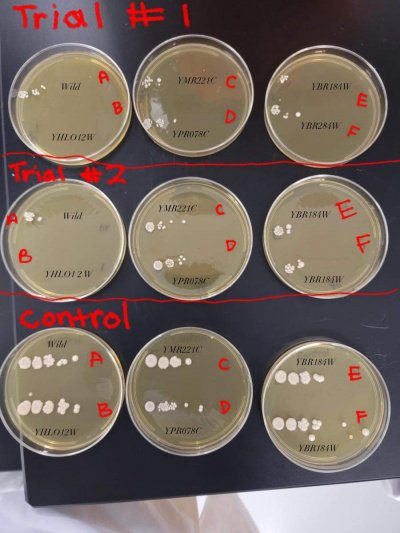
| + | [[Image: Heat Shock.jpeg|center|thumb|400px]] | ||
| + | YBR284W (F) | ||
| + | *This yeast strain had strong growth in the control group but was very stressed by the heat shock and showed less clusters/colonies on the heat shocked trial plates. The heat shocked trial plates showed a growth decrease by 4 times. | ||
| + | |||
| + | ===[[UW-Stout/Nitrogen_Starvation_FA21|Nitrogen Starvation]]=== | ||
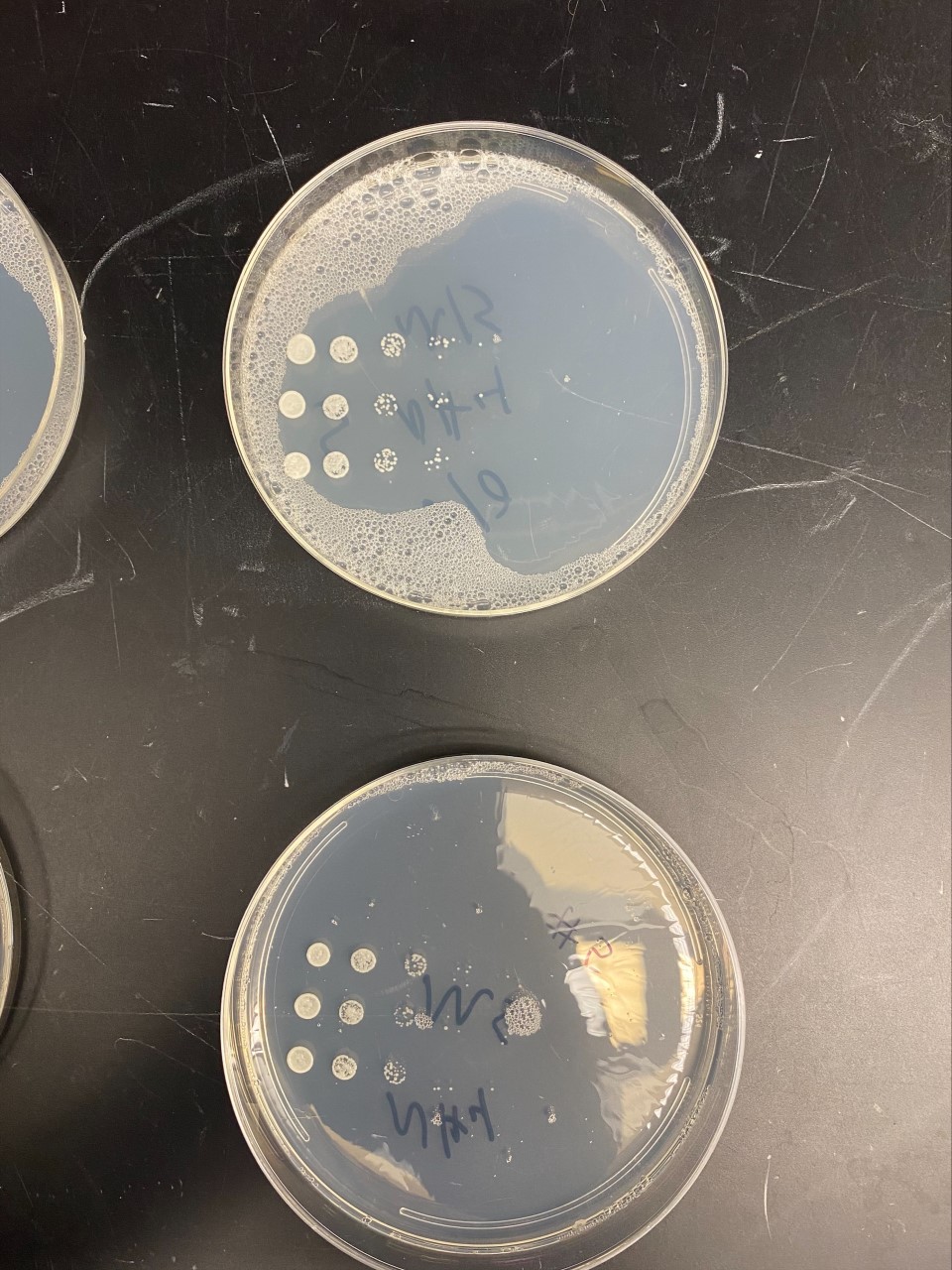
| + | [[File:YBR284W_Nitrogen21.jpg]] | ||
| + | |||
| + | *For the YBR284W gene, results showed that there was a slight increase in growth on the dish that did not contain NH₄ where there were more colonies of yeast. | ||
| + | |||
| + | ===[[UW-Stout/pH_FA21|pH]]=== | ||
| + | |||
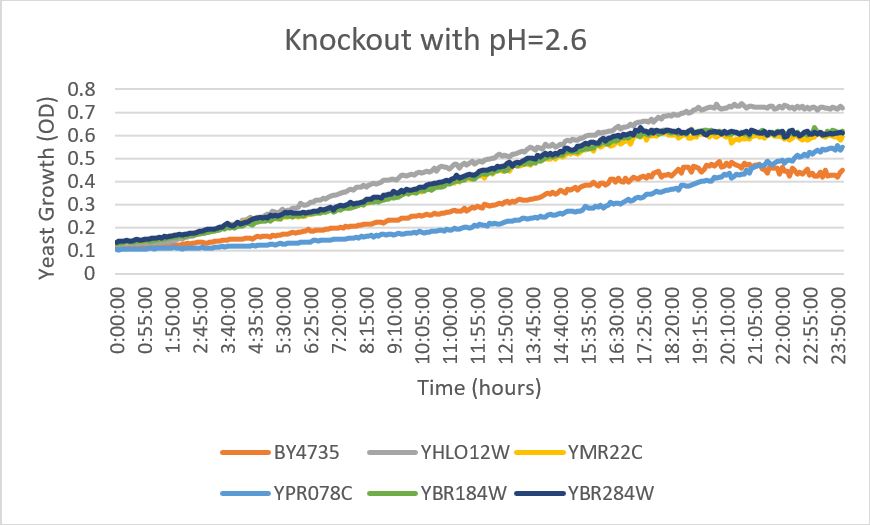
| + | [[File:pH_knockout.jpg]] | ||
| + | |||
| + | YBR284W | ||
| + | *When this strain was exposed to a citric acid buffer with a pH of 2.6, there did not appear to be a significant change in the growth rate of the strain | ||
| + | Protocol: | ||
| + | [[https://wiki.yeastgenome.org/index.php/UW-Stout/pH_FA21]] | ||
| + | |||
| + | ===[[UW-Stout/Bud Scars FA21]]=== | ||
| + | |||
| + | Cells grown with this gene (YBR284W) knocked out had an average of 1.18 bud scars per cell while the wild type (BY-RY4735) had 0.85 bud scars per cell. Of 114 YBR284W cells counted, 135 bud scars were seen. This indicated that the YBR284W gene slows reproduction rate and thus bud scarring in yeast cells. | ||
| + | |||
| + | |||
| + | ===[[UW-Stout/G418 FA21]]=== | ||
| + | |||
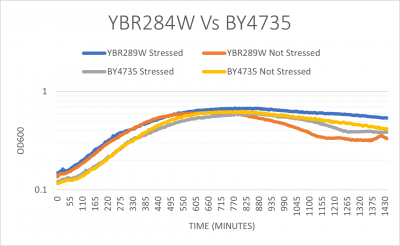
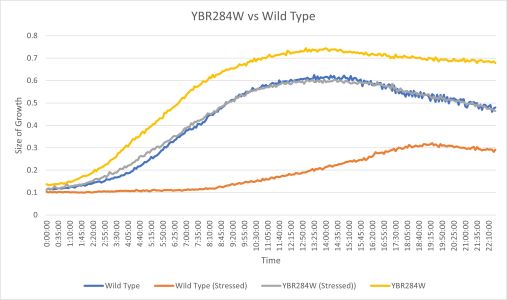
| + | [[File:YBR284W Vs BY4735 G418.png|left|thumb|400px]] | ||
| + | |||
| + | |||
| + | The YBR284W yeast strain doubling time without any stress was 174 minutes which is pretty similar to the wild type with 182 minutes. When both strains were stressed by G418 they YBR284W doubling time was 173 minutes and the wild type was 178 minutes. (These times are averaged between three trials) | ||
| + | Protocol: [https://wiki.yeastgenome.org/index.php/UW-Stout/G418_FA21] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
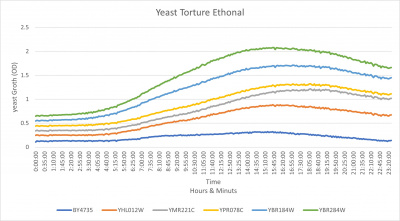
| + | ===[[UW-Stout/Ethanol FA21]]=== | ||
| + | [[File:Yeast ToutureV2.jpg|400px]] | ||
| + | * The Yeast Strain of YBR284W was not affected in any waty by the 18.5 % Ethanol it still has a steady growth curve | ||
| + | [[UW-Stout/Ethanol FA21]] | ||
| + | |||
| + | |||
| + | |||
| + | ===[[UW-Stout/Copper FA21]]=== | ||
| + | |||
| + | [[File:ybr2.jpg]] | ||
| + | |||
| + | The average doubling time for YBR284W stressed with our CuSO4 solution was 239 minutes, while unstressed was 207. The stressed version grew approximately 14% slower than the unstressed version. The unstressed version appears to have also grown larger overall. | ||
| + | |||
| Line 43: | Line 106: | ||
<protect> | <protect> | ||
| + | |||
==References== | ==References== | ||
<!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | <!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | ||
{{RefHelp}} | {{RefHelp}} | ||
</protect> | </protect> | ||
Latest revision as of 15:25, 21 December 2021
Share your knowledge...Edit this entry! <protect>
| Systematic name | YBR284W |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr II:771239..773632 |
| Primary SGDID | S000000488 |
Description of YBR284W: Protein of unknown function; some similarity to AMP deaminases but lacks key catalytic residues and does not rescue purine nucleotide metabolic defect of quadruple aah1 ade8 amd1 his1 mutant; null mutant exhibits longer telomeres, altered Ty mobility, decreased resistance to rapamycin and wortmannin, and synthetic phenotype with expression of alpha-synuclein; induced in response to hydrostatic pressure; not an essential gene[1][2][3][4][5][6][7]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
UW-Stout/Heat Shock FA21
YBR284W (F)
- This yeast strain had strong growth in the control group but was very stressed by the heat shock and showed less clusters/colonies on the heat shocked trial plates. The heat shocked trial plates showed a growth decrease by 4 times.
Nitrogen Starvation
- For the YBR284W gene, results showed that there was a slight increase in growth on the dish that did not contain NH₄ where there were more colonies of yeast.
pH
YBR284W
- When this strain was exposed to a citric acid buffer with a pH of 2.6, there did not appear to be a significant change in the growth rate of the strain
Protocol: [[1]]
UW-Stout/Bud Scars FA21
Cells grown with this gene (YBR284W) knocked out had an average of 1.18 bud scars per cell while the wild type (BY-RY4735) had 0.85 bud scars per cell. Of 114 YBR284W cells counted, 135 bud scars were seen. This indicated that the YBR284W gene slows reproduction rate and thus bud scarring in yeast cells.
UW-Stout/G418 FA21
The YBR284W yeast strain doubling time without any stress was 174 minutes which is pretty similar to the wild type with 182 minutes. When both strains were stressed by G418 they YBR284W doubling time was 173 minutes and the wild type was 178 minutes. (These times are averaged between three trials)
Protocol: [2]
UW-Stout/Ethanol FA21
- The Yeast Strain of YBR284W was not affected in any waty by the 18.5 % Ethanol it still has a steady growth curve
UW-Stout/Copper FA21
The average doubling time for YBR284W stressed with our CuSO4 solution was 239 minutes, while unstressed was 207. The stressed version grew approximately 14% slower than the unstressed version. The unstressed version appears to have also grown larger overall.
<protect>
References
See Help:References on how to add references
- ↑ Gatbonton T, et al. (2006) Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet 2(3):e35 SGD PMID 16552446
- ↑ Holmstrom K, et al. (1994) The sequence of a 32,420 bp segment located on the right arm of chromosome II from Saccharomyces cerevisiae. Yeast 10 Suppl A:S47-62 SGD PMID 8091861
- ↑ Iwahashi H, et al. (2003) Piezophysiology of genome wide gene expression levels in the yeast Saccharomyces cerevisiae. Extremophiles 7(4):291-8 SGD PMID 12910389
- ↑ Maxwell PH and Curcio MJ (2007) Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for regulation of mammalian retroviruses. Eukaryot Cell 6(7):1069-80 SGD PMID 17496126
- ↑ Saint-Marc C, et al. (2009) Phenotypic consequences of purine nucleotide imbalance in Saccharomyces cerevisiae. Genetics 183(2):529-38, 1SI-7SI SGD PMID 19635936
- ↑ Willingham S, et al. (2003) Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science 302(5651):1769-72 SGD PMID 14657499
- ↑ Xie MW, et al. (2005) Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method. Proc Natl Acad Sci U S A 102(20):7215-20 SGD PMID 15883373
See Help:Categories on how to add the wiki page for this gene to a Category </protect>