Difference between revisions of "UW-Stout/UV Light SP22"
(Created page with "To knock out genes in S. cerevisiae, we used a Cas9-assisted homologous recombination approach: * Build an S. cerevisiae shuttle vector expressing Cas9 and a guide RNA targeti...") |
|||
| Line 1: | Line 1: | ||
| − | + | ==Materials== | |
| − | * | + | *Cell 60 mm Culture Dish, containing 6 ml of agar |
| − | * | + | *Phosphate Buffered Saline (PBS) |
| − | * | + | *Glycerol stocks of yeast strains |
| − | * | + | *15ml centrifuge tubes |
| − | == | + | ==Equipment== |
| + | *Incubator set to 30°C. | ||
| + | *Bachur & Associates Sanata Clara, CA 95050 Model LS-100-3 UV Light Exposure System | ||
| + | *P1000 and P10 Micro-pipettes | ||
| − | * | + | ==Calibration Protocol== |
| − | + | *Wear rubber gloves when handling yeast samples, avoid direct exposure to UV light and wear safety glasses if needed. | |
| + | #Fill a 15ml centrifuge tube with 9,990ul of PBS. | ||
| + | #Vortex yeast stock to resuspend the yeast cells. | ||
| + | #Pipette 10ul of wild yeast stock into the 9,990ul of PBS to create a dilution containing 2 yeast cells per microliter. | ||
| + | #Vortex dilution and prepare 7 plates with 50ul of the wild yeast dilution for about 100 yeast cells per plate. | ||
| + | #Label each plate individually 0, 500, 600, 700, 800, 900 and 1000 for the number of seconds each plate will be exposed to the UV light. | ||
| + | #Set up the UV light exposure system: | ||
| + | ##400 watts | ||
| + | ##desired time increment | ||
| + | #Run yeast plates (without plate top) under the UV light for their respective times in seconds. | ||
| + | #Place plates upside down in dark incubator set to 30°C for 48 hours. | ||
| + | #Count number of colonies on each plate using the 0 second plate as your control to compare to. Based on how many colonies there are on each plate, determine the time frame that killed roughly 50% of the yeast cells. | ||
| − | == | + | ==Calibration Results== |
| − | + | [[Image:4A1B2FAF-1C83-4A7A-AE89-FFC5BFBE7D35.jpeg|center|thumb|400px|CalibrationResults1]] | |
| − | + | [[Image:C5D74C8D-33BF-426D-9E2F-C0E9385AE258.jpeg|center|thumb|400px|CalibrationResults2]] | |
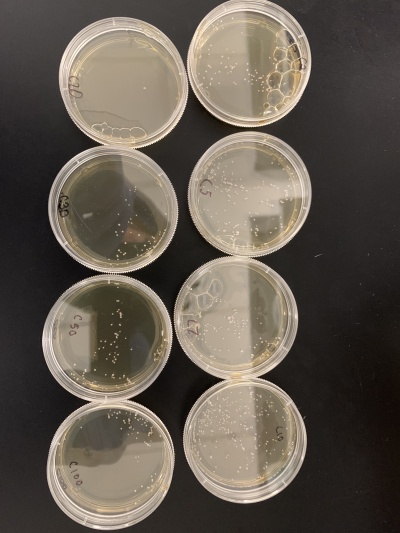
| − | + | *For calibration trial 1 we ran 14 plates at varying times of 2, 5, 7, 10, 20, 30, 50, 100, 150, 200, 250, 300, 400 and 500 seconds. After giving the yeast time to grow we counted the colonies in each plate. | |
| − | + | *'''Results''':2sec=139 colonies, 5sec=150 colonies, 7sec=147 colonies, 10sec=296 colonies, 20sec=16 colonies, 30sec=82 colonies, 50sec=92 colonies, 100sec=125 colonies, 150sec=289 colonies, 200sec=203 colonies, 250sec=29 colonies, 300sec=214 colonies, 400sec=176 colonies, 500sec=200 colonies. | |
| − | + | *'''Interpretation''': Even at 500 seconds there were still a lot of yeast colonies meaning 500 seconds likely wasn't long enough to stress the yeast cells as desired. There were a couple outliers that had very few colonies despite not being under the UV light for very long. This was likely because the plates didn't have the desired number of cells since it is impossible to tell exactly how many cells are on the plate until they've had time to grow. | |
| − | + | [[Image:5517307E-5966-453B-98F0-1EBB618F5ABB.jpeg|center|thumb|400px|CalibrationResults3]] | |
| − | + | [[Image:FA26DC6B-6AFA-4ADD-80D9-67441E11BE3D.jpeg|center|thumb|400px|CalibrationResults4]] | |
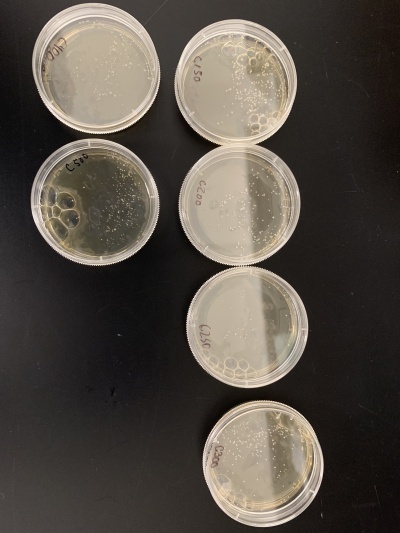
| − | + | *For calibration trial 2 we ran 6 more plates at varying times of 500, 600, 700, 800, 900 and 1000. We counted the colonies after giving the yeast time to grow. | |
| − | + | *'''Results''':500sec=123 colonies, 600sec=29 colonies, 700sec=11 colonies, 800sec=6 colonies, 900=1 colony, 1000sec=1 colony. | |
| + | *'''Interpretation''': We noticed that like before 500 seconds still had a lot of yeast colonies however 600 and 700 seconds had closer to the desired amount of yeast colonies. 800, 900 and 1000 seconds had too few yeast cells after exposure meaning these were too long to be used. With this information we decided to use 600 seconds of UV exposure for our knock-out experiments. | ||
| + | ==Knock-out Protocol== | ||
| + | *Wear rubber gloves when handling yeast samples, avoid direct exposure to UV light and wear safety glasses if needed. | ||
| + | #Fill 7 15ml centrifuge tube with 9,990ul of PBS. | ||
| + | #Vortex each yeast stock to resuspend the yeast cells. (wild and 6 knock-out strains) | ||
| + | #Pipette 10ul of each respective yeast stock into one of the centrifuge tubes containing 9,990ul of PBS to create a dilution containing 2 yeast cells per microliter for each strain. Make sure to label each tube so the strains don't get mixed up. | ||
| + | #Vortex each dilution and prepare 14 plates total, two for each strain of yeast with 50ul of the yeast dilution for about 100 yeast cells per plate. | ||
| + | #Label 7 plates with 0 and the other 7 plates with 600 for the number of seconds each plate will be exposed to the UV light. Also label what strain of yeast in in each plate, there should be a 0 plate and 600 plate for each strain. | ||
| + | #Set up the UV light exposure system: | ||
| + | ##400 watts | ||
| + | ##desired time increment | ||
| + | #Run yeast plates (without plate top) under the UV light for their respective times in seconds. | ||
| + | #Place plates upside down in dark incubator set to 30°C for 48 hours. | ||
| + | #Count number of colonies on each plate using the 0 second plate as your control to compare to. Based on how many colonies there are on each plate, determine if the knocked-out gene of the yeast had any affect on the survival of the yeast cells (improved/reduced survival). | ||
| + | #Repeat the procedure above as needed to acquire the data needed. | ||
| − | == | + | ==Knock-out Results== |
| − | + | *Knock-out strain 1:[[YOR387C]] | |
| − | + | *Knock-out strain 2:[[YKL121W]] | |
| − | + | *Knock-out strain 3:[[YBR225W]] | |
| − | + | *Knock-out strain 4:[[YNL018C]] | |
| − | == | + | *Knock-out strain 5:[[YLR426W]] |
| − | + | *Knock-out strain 6:[[YGL138C]] | |
| − | + | ====Wild type==== | |
| − | + | [[Image:FD478531-2B52-4E05-9D2B-9D9DCBC79219.jpeg|center|thumb|400px|Wild strain]] | |
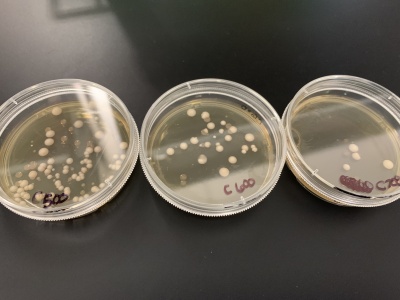
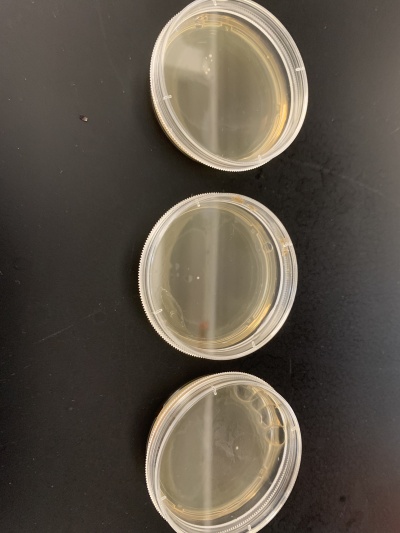
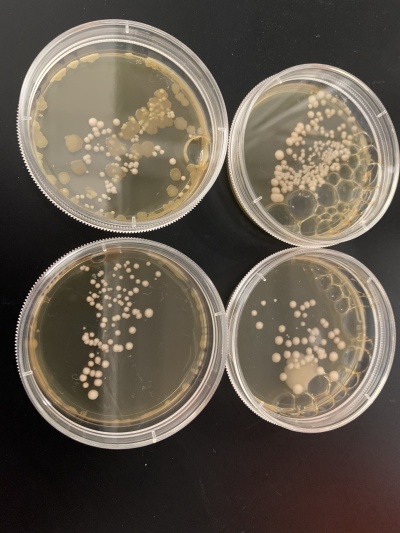
| − | + | '''Results''': | |
| − | + | *Experiment 1:0sec=24 colonies, 600sec=58 colonies. | |
| − | + | *Experiment 2:0sec=95 colonies, 600sec=92 colonies. | |
| − | + | '''Interpretation''': The two 0sec plates are on top and the 600sec plates are on the bottom. I believe something went wrong with the wild strain run. The 0sec plat had fewer than the plate exposed to the UV light for 600sec and the second run didn't seem to be affected much by the UV light. My guess is that the number of cells each plate had in the beginning was significantly different making it hard for this data to be used reliably. | |
Revision as of 12:26, 21 April 2022
Contents
Materials
- Cell 60 mm Culture Dish, containing 6 ml of agar
- Phosphate Buffered Saline (PBS)
- Glycerol stocks of yeast strains
- 15ml centrifuge tubes
Equipment
- Incubator set to 30°C.
- Bachur & Associates Sanata Clara, CA 95050 Model LS-100-3 UV Light Exposure System
- P1000 and P10 Micro-pipettes
Calibration Protocol
- Wear rubber gloves when handling yeast samples, avoid direct exposure to UV light and wear safety glasses if needed.
- Fill a 15ml centrifuge tube with 9,990ul of PBS.
- Vortex yeast stock to resuspend the yeast cells.
- Pipette 10ul of wild yeast stock into the 9,990ul of PBS to create a dilution containing 2 yeast cells per microliter.
- Vortex dilution and prepare 7 plates with 50ul of the wild yeast dilution for about 100 yeast cells per plate.
- Label each plate individually 0, 500, 600, 700, 800, 900 and 1000 for the number of seconds each plate will be exposed to the UV light.
- Set up the UV light exposure system:
- 400 watts
- desired time increment
- Run yeast plates (without plate top) under the UV light for their respective times in seconds.
- Place plates upside down in dark incubator set to 30°C for 48 hours.
- Count number of colonies on each plate using the 0 second plate as your control to compare to. Based on how many colonies there are on each plate, determine the time frame that killed roughly 50% of the yeast cells.
Calibration Results
- For calibration trial 1 we ran 14 plates at varying times of 2, 5, 7, 10, 20, 30, 50, 100, 150, 200, 250, 300, 400 and 500 seconds. After giving the yeast time to grow we counted the colonies in each plate.
- Results:2sec=139 colonies, 5sec=150 colonies, 7sec=147 colonies, 10sec=296 colonies, 20sec=16 colonies, 30sec=82 colonies, 50sec=92 colonies, 100sec=125 colonies, 150sec=289 colonies, 200sec=203 colonies, 250sec=29 colonies, 300sec=214 colonies, 400sec=176 colonies, 500sec=200 colonies.
- Interpretation: Even at 500 seconds there were still a lot of yeast colonies meaning 500 seconds likely wasn't long enough to stress the yeast cells as desired. There were a couple outliers that had very few colonies despite not being under the UV light for very long. This was likely because the plates didn't have the desired number of cells since it is impossible to tell exactly how many cells are on the plate until they've had time to grow.
- For calibration trial 2 we ran 6 more plates at varying times of 500, 600, 700, 800, 900 and 1000. We counted the colonies after giving the yeast time to grow.
- Results:500sec=123 colonies, 600sec=29 colonies, 700sec=11 colonies, 800sec=6 colonies, 900=1 colony, 1000sec=1 colony.
- Interpretation: We noticed that like before 500 seconds still had a lot of yeast colonies however 600 and 700 seconds had closer to the desired amount of yeast colonies. 800, 900 and 1000 seconds had too few yeast cells after exposure meaning these were too long to be used. With this information we decided to use 600 seconds of UV exposure for our knock-out experiments.
Knock-out Protocol
- Wear rubber gloves when handling yeast samples, avoid direct exposure to UV light and wear safety glasses if needed.
- Fill 7 15ml centrifuge tube with 9,990ul of PBS.
- Vortex each yeast stock to resuspend the yeast cells. (wild and 6 knock-out strains)
- Pipette 10ul of each respective yeast stock into one of the centrifuge tubes containing 9,990ul of PBS to create a dilution containing 2 yeast cells per microliter for each strain. Make sure to label each tube so the strains don't get mixed up.
- Vortex each dilution and prepare 14 plates total, two for each strain of yeast with 50ul of the yeast dilution for about 100 yeast cells per plate.
- Label 7 plates with 0 and the other 7 plates with 600 for the number of seconds each plate will be exposed to the UV light. Also label what strain of yeast in in each plate, there should be a 0 plate and 600 plate for each strain.
- Set up the UV light exposure system:
- 400 watts
- desired time increment
- Run yeast plates (without plate top) under the UV light for their respective times in seconds.
- Place plates upside down in dark incubator set to 30°C for 48 hours.
- Count number of colonies on each plate using the 0 second plate as your control to compare to. Based on how many colonies there are on each plate, determine if the knocked-out gene of the yeast had any affect on the survival of the yeast cells (improved/reduced survival).
- Repeat the procedure above as needed to acquire the data needed.
Knock-out Results
- Knock-out strain 1:YOR387C
- Knock-out strain 2:YKL121W
- Knock-out strain 3:YBR225W
- Knock-out strain 4:YNL018C
- Knock-out strain 5:YLR426W
- Knock-out strain 6:YGL138C
Wild type
Results:
- Experiment 1:0sec=24 colonies, 600sec=58 colonies.
- Experiment 2:0sec=95 colonies, 600sec=92 colonies.
Interpretation: The two 0sec plates are on top and the 600sec plates are on the bottom. I believe something went wrong with the wild strain run. The 0sec plat had fewer than the plate exposed to the UV light for 600sec and the second run didn't seem to be affected much by the UV light. My guess is that the number of cells each plate had in the beginning was significantly different making it hard for this data to be used reliably.