YLR426W
Share your knowledge...Edit this entry! <protect>
| Systematic name | YLR426W |
| Gene name | TDA5 |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr XII:987062..988113 |
| Primary SGDID | S000004418 |
Description of YLR426W: Putative protein of unknown function; detected in highly purified mitochondria in high-throughput studies; proposed to be involved in resistance to mechlorethamine and streptozotocin; null mutant sensitive to expression of top1-T722A allele[1][2][3][4]
</protect>
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
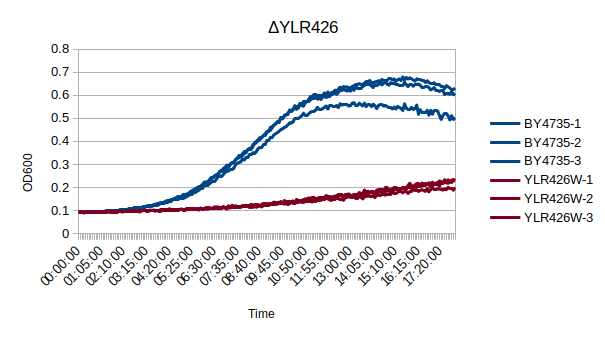
Growth Curve
In a BY4735 background, knocking out YLR426W seems to have a substantial effect on the strain's growth in complete media. In this assay, the BY4735 strain's doubling time was 162 minutes, while the YLR426W knock-out strain's doubling time was 660 minutes. (These doubling times are the means of three experiments.)
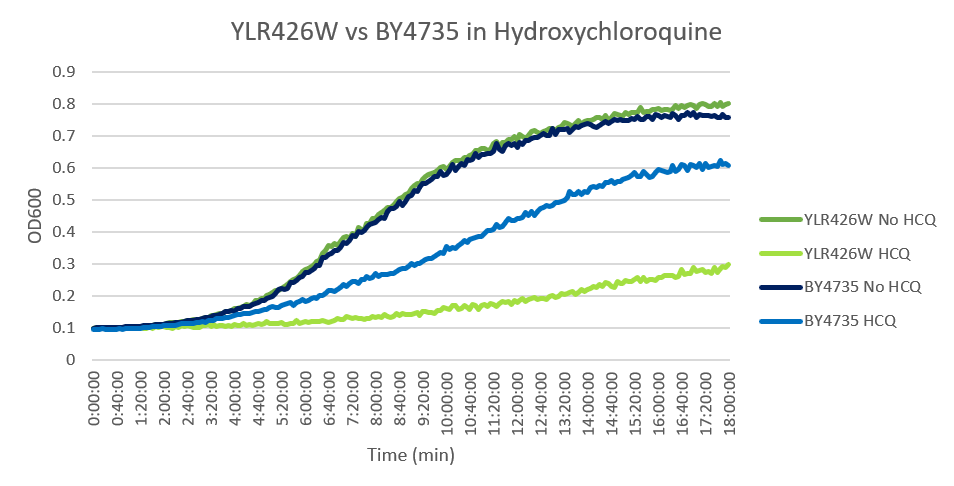
Hydroxychloroquine
HCQ refers to hydroxychloroquine
Under normal conditions, the BY4735 doubling time is 144 minutes, while that of YLR426W is 139 minutes. This proves that both of these strains grow at a similar rate. When grown in an environment with hydroxychloroquine, YLR426W doubled in 505 minutes, while BY4735 doubled in 276 minutes. Both strains are inhibited by hydroxychloroquine, but YLR426W growth is staggered much more. This is a possible indicator that the knocked-out gene was involved in cellular protection from hydroxychloroquine. (These doubling times and curves are the means of three experiments.)
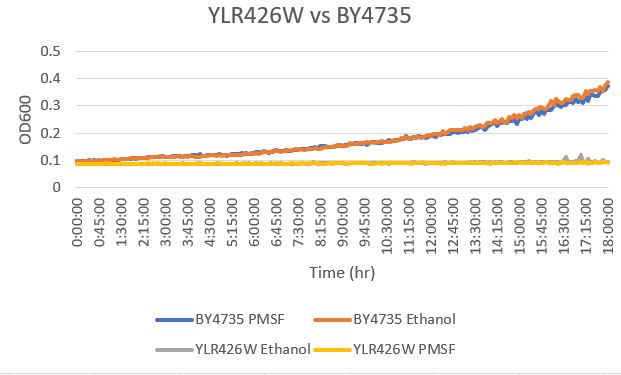
Phenylmethylsulphonyl Fluoride
The YLR426W yeast gene was effected drastically by the addition of PMSF. With a doubling time of 96,408 minutes with the PMSF present, it did far worse than the BY4735 strain. This is likely due to a critical change in the yeast's ability to handle chemicals like PMSF that effectively break the cell down through cell lysis. The BY4735 had a doubling time of 382 minutes. The growth of the YLR426W strain was essentially nonexistent in this case. The ethanol growth was also very low at a doubling time of 95,987 minutes on average. The doubling time of the BY4735 strain was 442 minutes.
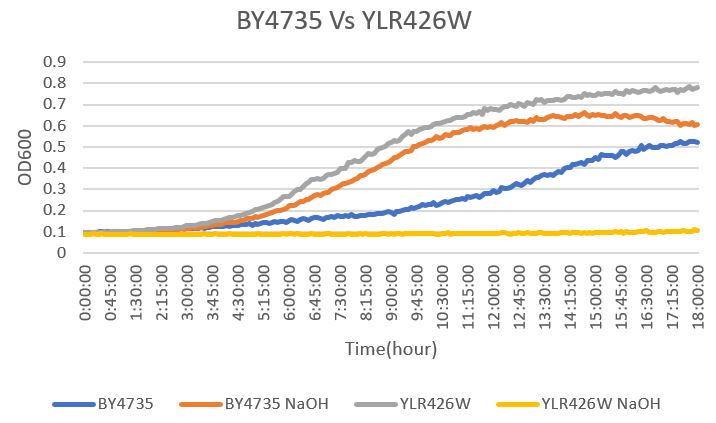
=Sodium Hydroxide
The YLR426W yeast gene was effected drastically by NaOH. With a doubling time of 714 minutes with the NaOH present, it did far worse than the BY4735 strain. This is likely due to a critical change in the yeast's ability to handle chemicals like NaOH that effectively break the cell down through cell lysis. The BY4735 had a doubling time of 545 minutes. The growth of the YLR426W strain was essentially nonexistent in this case. The doubling time with water for YLR426W yeast gene was 2482 minutes and the BY4735 is 809 minutes.
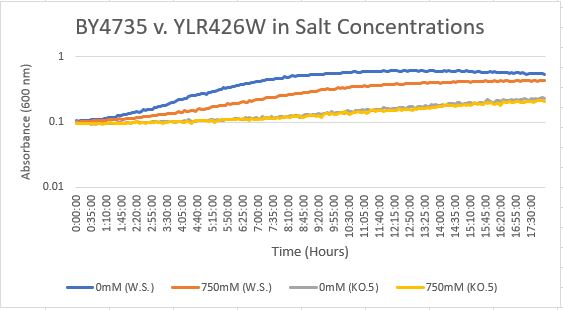
Salt Concentration (NaCl)
0mM NaCl conc. BY4735 strain's doubling time: 169 minutes
0mM NaCl conc. YLR426W strain's doubling time: 481 minutes
750mM NaCl conc. BY4735 strain's doubling time: 294 minutes
750mM NaCl conc. YLR426W strain's doubling time: 909 minutes
The graph above shows the growth rate for the previously listed strains and the relative level of NaCl concentration. Knocking out the gene negatively and severely impacted the growth rate. Comparing the YLR426W doubling times between 0mM and 750mM NaCl, there is a tremendous effect on the growth rate. This effect negatively influences the knockout strain's (YLR426W) growth rate.
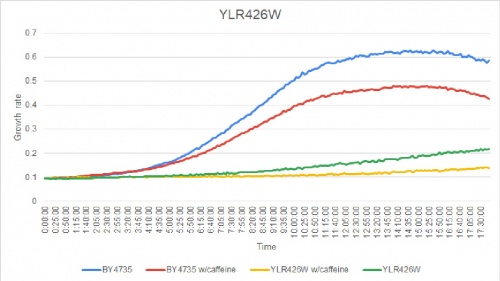
Caffeine Group 1
YLR426W was greatly effected by caffeine. The doubling times are as follows:
Doubling time of YLR426W with caffeine: 1267 minutes
Doubling time of YLR426W without caffeine: 294 minutes
Doubling time of BY4735 with caffeine: 138 minutes
Doubling time of BY4735 without caffeine: 187 minutes
The strain's growth rate had slightly slowed when caffeine was added. The growth rate of this knockout strain already was slower than others to begin with.
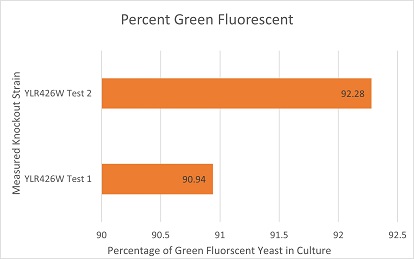
Competative Co-culture
These are the results of a competitive co-culture protocol. The knockout strain was grown in a culture to test fitness against a green, fluorescent wild-type strain. The percentage of analyzed cells in the culture measured was those displaying green fluorescence. In theory, this should mean if a knockout has reduced the fitness of a strain of yeast, the fluorescent wild-type strain would have a higher percentage than the knockout strain. If the knockout does not decrease fitness, they would be roughly equal. This may not be so in results. Sources of error may include contamination and human error.
The results here are expected in theory. The knockout of YLR426W suggests the knockout strain had a major disadvantage against the wild-type fluorescent strain. There are also two things worth mentioning. One: The knockout strain might display a slight green fluorescence but a technical problems over the short time frame of experimentation made this hard to confirm. If this is true, that would skew the results somewhat. Two: It was noted throughout experimentation that the knockout strain grew incredibly slowly on its own in a liquid or plate culture.
Ammonium Sulfate
YLR426W gene does not seem to be affected by nitrogen starvation. We know this because of the doubling times. With this gene knocked out it grows 9.6% slower than that of the wild which grows 17.8% slower. This is not a significant difference in the scope of things. Though it is to note that this gene when knocked out causes significantly slower growth rates than that of its peers. The comparison is YLR426W at 11 hours and its peers 2-4 hours.
<protect>
UV Light
Results:
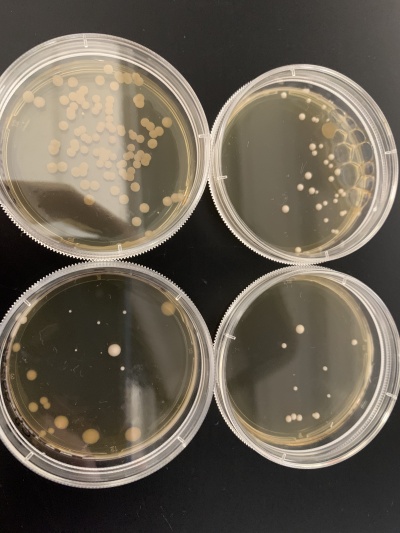
- Experiment 1:0sec=37 colonies, 600sec=11 colonies.
- Experiment 2:0sec=0 colonies, 600sec=7 colonies.
Interpretation: The two 0sec plates are on top and the 600sec plates are on the bottom. None of the plates for this yeast strain seemed to have many colonies with the 0sec plate from experiment 2 having no colonies at all and only contamination. Because of this, it's hard to say if the knocked-out gene is related to the UV light since we can only really use the first experiment (right). Another experiment would need to be done to have significant data. My guess is that the 0sec plate from experiment 2 had no yeast cells when the plate was prepared since the UV light can't be blamed.
Calcofluor White
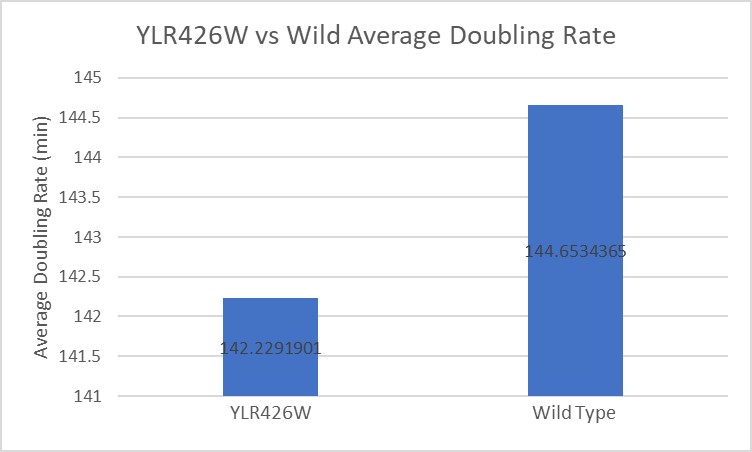
From this data we can determine that there was minimal growth inhibition on this strain of yeast. The modified yeast had a doubling rate that was actually about two minutes faster than the wild type on average, and when compared to the effect on other yeast strains this is would lead us to believe there was no inhibiting effect on the modified yeast.
Heat Shock
The YLR426W yeast strain was effected the most by heat shocking than any other gene tested in this protocol/experiment. The controlled plate for this gene grew a very thick blanket of DNA colonies (comparable to the YBR225W strain). As seen in the picture, the controlled plate (labeled "X") was covered with cells, while the two trial plates had very little growth, if anything at all. This specific yeast strain was very sensitive to any sort of heat. Most other genes had at least a little growth to a decent amount, but this one failed to handle the heat shock.
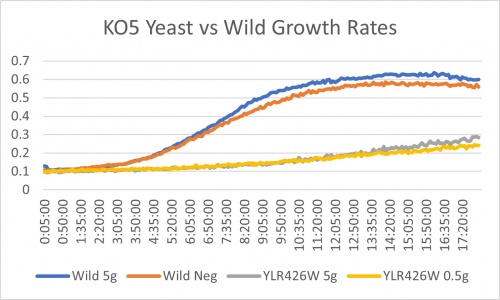
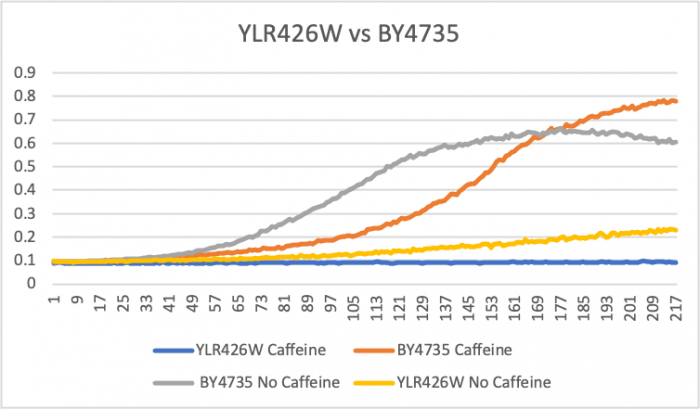
Caffeine Group 2
Doubling Time at 0.25 concentration
Wild Type Td = (146-66) x ln(2)/ln(.6/.2) = 88 minutes; Wild Type no caffeine Td= (113-33) x ln(2)/ln(.5/.1)=34 minutes; YLR426W no caffeine Td= (193-73) x ln(2)/ln(.2/.1)= 120 minutes; YLR426W Td = (217-1) x ln(2)/ln(0.1/0.09) = 1421 minutes These doubling times show that the 0.25 mM concentration in YLR426W slowed down the rate of growth. The caffeine was able to slow down metabolic activity. Caffeine may effect the function of the Rad52 protein that aids in DNA double-stranded breaks and maintaining genome stability. The caffeine may also aid in the process of the cell protecting itself from reacting oxygen species. Since the concentration is small in volume, apoptosis was not induced for cell function.
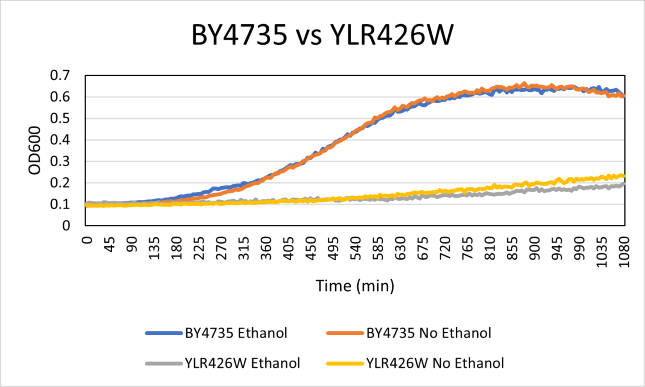
Ethanol Sensitivity
The YLR426W yeast strain was effected drastically by the addition of ethanol compared to the BY4735 strain. With a doubling time of 665 minutes with ethanol absent. The BY4735 had a doubling time of 182 minutes. The growth of the YLR426W strain was much slower. The ethanol growth was also very low at a doubling time of 777 minutes on average. The doubling time of the BY4735 strain was 218 minutes with ethanol.
[[1]]
Hydroxurea
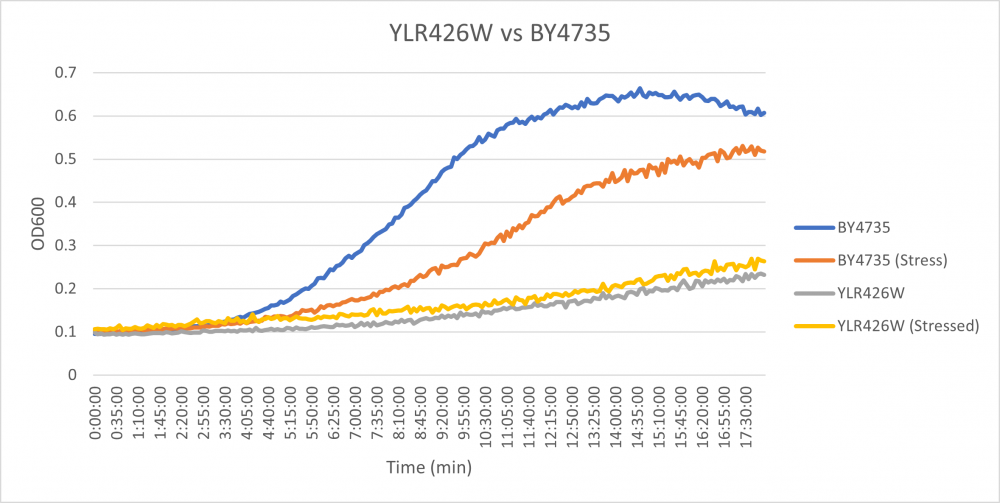
At first glance, the YLR426W strain seems to have a large difference in growth compared to the BY4735 strain. The YLR426W strain when exposed to hydroxyurea had an average doubling time of 946 minutes, while the BY4735 strain when stressed had a doubling time of 278 minutes. The YLR426W strain without stress had an average doubling time of 710 minutes, while the BY4735 strain unstressed had an average doubling time of 182 minutes. Through analysis, it seems the YLR426W strain just seems to grow slower than other strains.
References
See Help:References on how to add references
- ↑ Lee W, et al. (2005) Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet 1(2):e24 SGD PMID 16121259
- ↑ Reid RJ, et al. (2011) Selective ploidy ablation, a high-throughput plasmid transfer protocol, identifies new genes affecting topoisomerase I-induced DNA damage. Genome Res 21(3):477-86 SGD PMID 21173034
- ↑ Reinders J, et al. (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5(7):1543-54 SGD PMID 16823961
- ↑ Sickmann A, et al. (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A 100(23):13207-12 SGD PMID 14576278
See Help:Categories on how to add the wiki page for this gene to a Category </protect>