YDL109C
Share your knowledge...Edit this entry! <protect>
| Systematic name | YDL109C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr IV:267201..265258 |
| Primary SGDID | S000002267 |
Description of YDL109C: Putative lipase; involved in lipid metabolism; YDL109C is not an essential gene[1]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
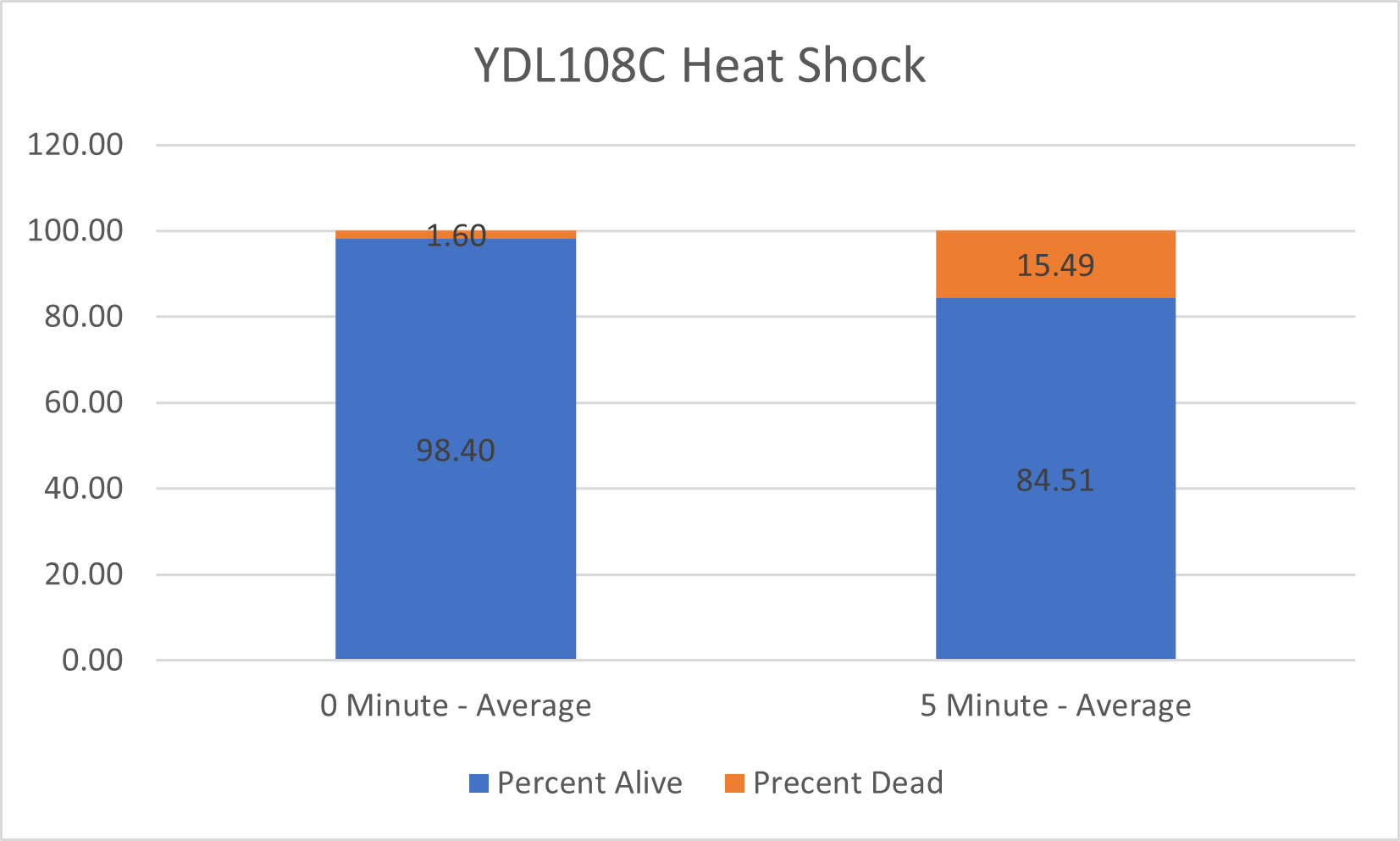
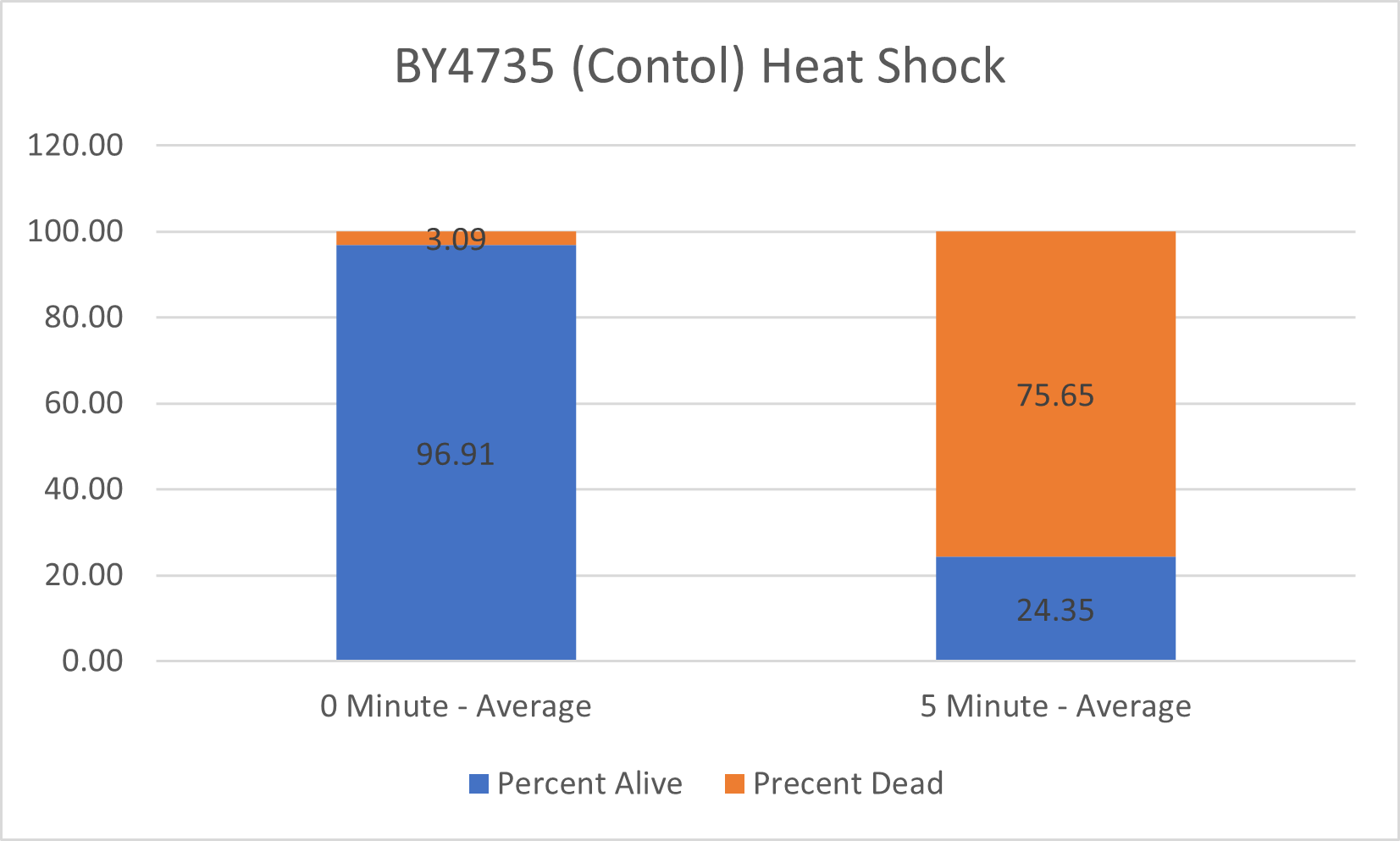
UW-Stout/Heat Shock SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to heat shock.
Results
Interpretation
From the data gathered, there was a large positive effect to knocking out this gene when it came to the yeast's ability to hold up to heat shock. The modified yeast cells were 350% more resistant to heat shock than the control.
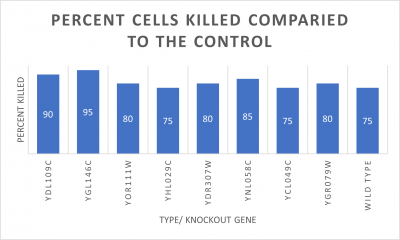
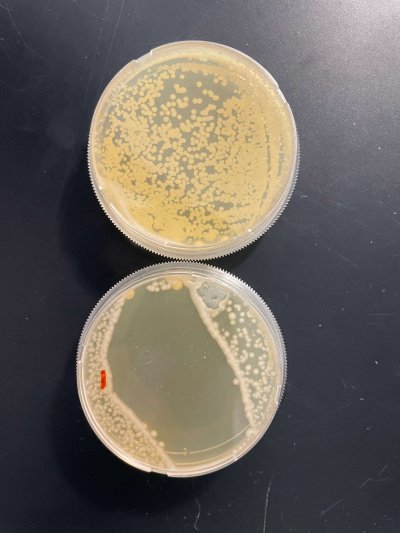
UW-Stout/UV Light SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested under a UV Light using this protocol.
RESULTS
INTERPERTATION
In the graph and photos above you can see that exposing this gene to 600 seconds of 400 Watt UV Light killed approximately 90% of yeast cell cultures, in comparison to its control counterpart which was the the same gene and amount of cells, just was not exposed to UV Light.
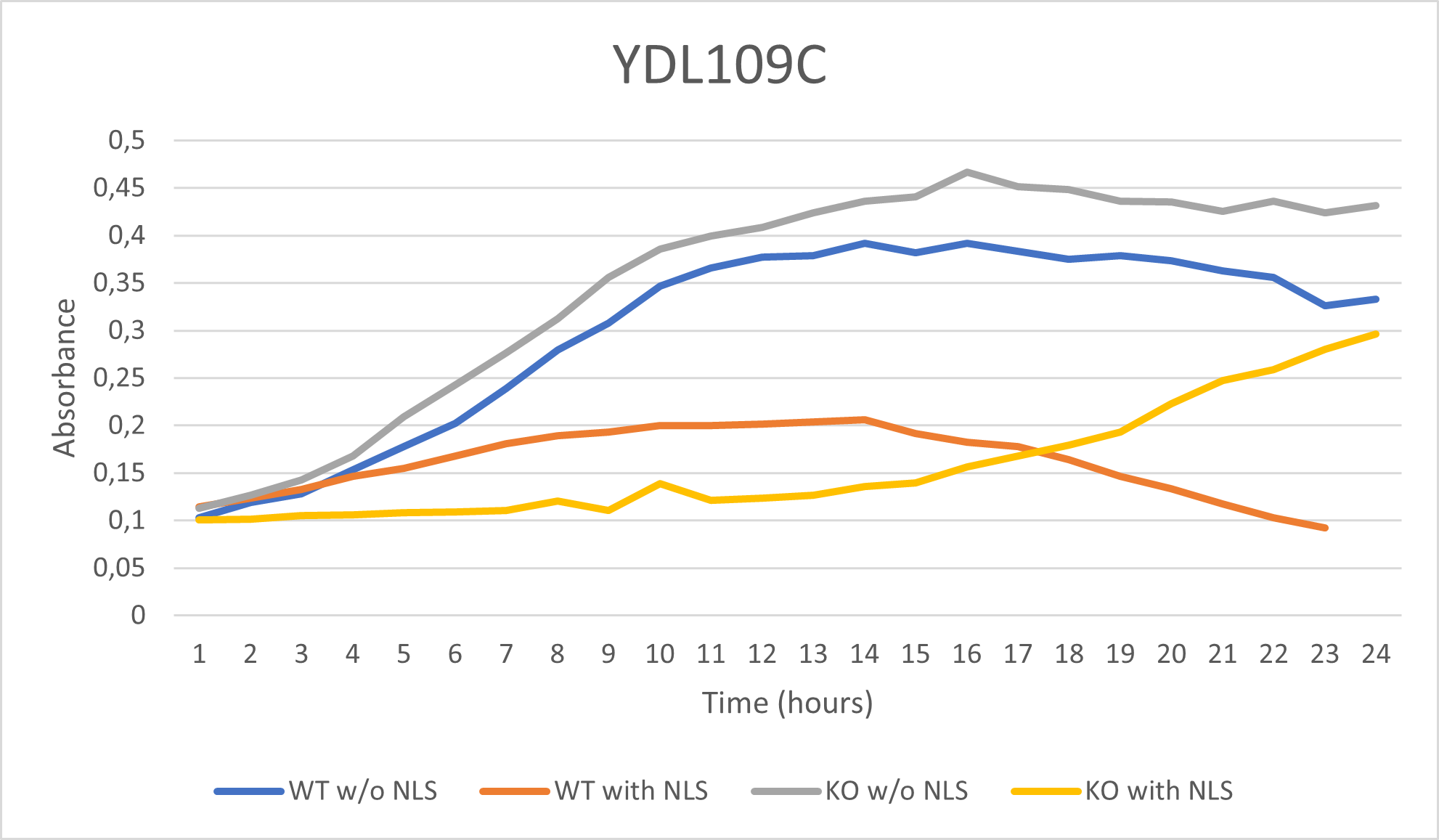
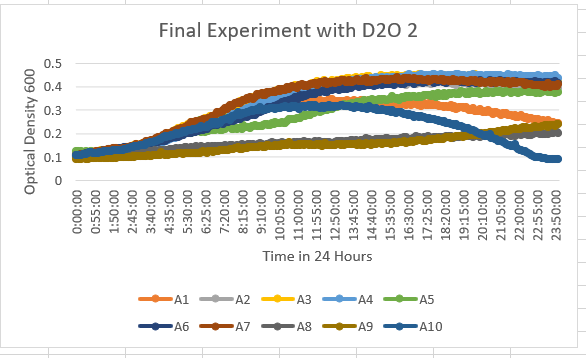
UW-Stout/D2O SP22
The YDL109C (A1) Knock Out Yeast Strain was a little more in the middle spectrum of results with being a little more resistant to the 35% dilution of D2O. The range it stays in as a maximum OD 600 is 0.3-0.4. With the growth curve decaying at the end at about 16 hours.
DNA and RNA Details
Other DNA and RNA Details
Other Topic: expression
Specifically higher expression in phosphorus limited chemostat cultures versus phosphorus excess. [2] [3]
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
<protect>
References
See Help:References on how to add references
- ↑ Daum G, et al. (1999) Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast 15(7):601-14 SGD PMID 10341423
- ↑ Boer VM, et al. (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278(5):3265-74 SGD PMID 12414795
- ↑ submitted by Viktor Boer on 2003-07-25
See Help:Categories on how to add the wiki page for this gene to a Category </protect>