Difference between revisions of "YDL109C"
(→UW-Stout/UV Light SP22) |
(→UW-Stout/UV Light SP22) |
||
| Line 37: | Line 37: | ||
[[File:ASHTONMIKOLOSKIFinal Project Graph.png|400px|]] | [[File:ASHTONMIKOLOSKIFinal Project Graph.png|400px|]] | ||
| − | [[File:AshtonGene1.jpg| | + | [[File:AshtonGene1.jpg|350px|]] |
'''INTERPERTATION''' | '''INTERPERTATION''' | ||
| − | In the graph and photos above | + | In the graph and photos above, exposing this gene to 600 seconds of 400 Watt UV Light killed approximately 90% of yeast cell cultures, compared to its control counterpart, which was the same gene and amount of cells, just was not exposed to UV Light. |
===[[UW-Stout/D2O SP22]]=== | ===[[UW-Stout/D2O SP22]]=== | ||
Revision as of 12:39, 3 May 2022
Share your knowledge...Edit this entry! <protect>
| Systematic name | YDL109C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr IV:267201..265258 |
| Primary SGDID | S000002267 |
Description of YDL109C: Putative lipase; involved in lipid metabolism; YDL109C is not an essential gene[1]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
UW-Stout/UV Light SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested under a UV Light using this protocol.
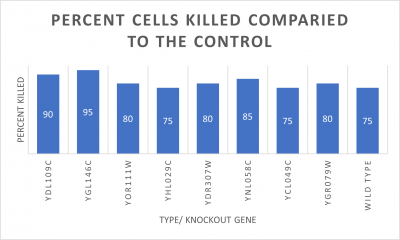
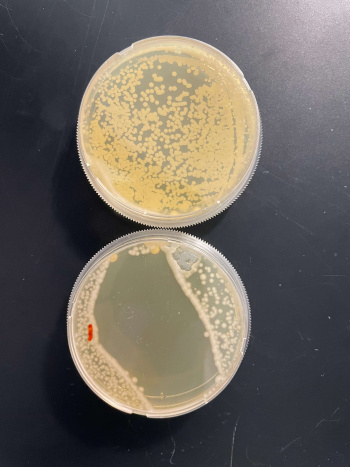
RESULTS
INTERPERTATION
In the graph and photos above, exposing this gene to 600 seconds of 400 Watt UV Light killed approximately 90% of yeast cell cultures, compared to its control counterpart, which was the same gene and amount of cells, just was not exposed to UV Light.
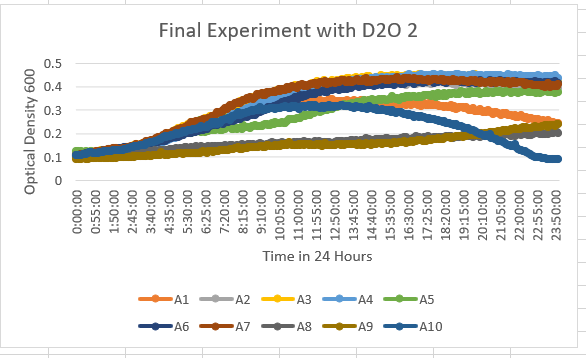
UW-Stout/D2O SP22
The YDL109C (A1) Knock Out Yeast Strain was a little more in the middle spectrum of results with being a little more resistant to the 35% dilution of D2O. The range it stays in as a maximum OD 600 is 0.3-0.4. With the growth curve decaying at the end at about 16 hours.
DNA and RNA Details
Other DNA and RNA Details
Other Topic: expression
Specifically higher expression in phosphorus limited chemostat cultures versus phosphorus excess. [2] [3]
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
<protect>
References
See Help:References on how to add references
- ↑ Daum G, et al. (1999) Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast 15(7):601-14 SGD PMID 10341423
- ↑ Boer VM, et al. (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278(5):3265-74 SGD PMID 12414795
- ↑ submitted by Viktor Boer on 2003-07-25
See Help:Categories on how to add the wiki page for this gene to a Category </protect>