YGL235W
Share your knowledge...Edit this entry!
| Systematic name | YGL235W |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr VII:55279..55815 |
| Primary SGDID | S000003204 |
Description of YGL235W: Putative protein of unknown function; potential Cdc28p substrate; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2[1][2]
Contents
- 1 Community Commentary
- 2 References
Community Commentary
About Community Commentary. Please share your knowledge!
Protein Details
Protein Modification
Modification(s): Phosphorylation
Identified as an efficient substrate of Clb2-Cdk1-as1 in a screen of a proteomic GST-fusion library. [2] [3]
UW-Stout/Sensitivity To Nitrogen Starvation
Knocking out the YGL235Wseems to have no effect on growth after incubating for 5 days on the Nitrogen omitted media.
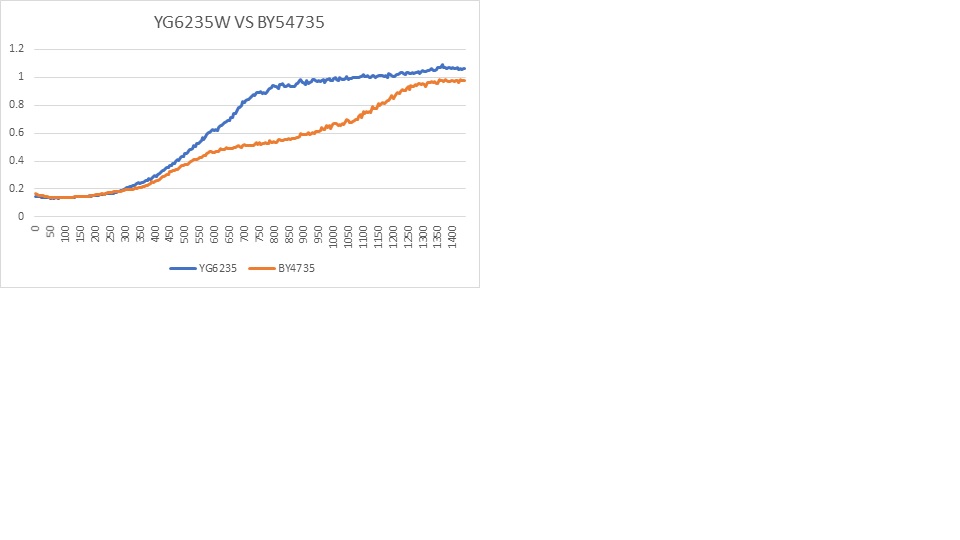
Ultraviolet Sensitivity
YGL235W is more sensitive than BY4735(wild) given that there is less colonizes present when under the same stress.
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
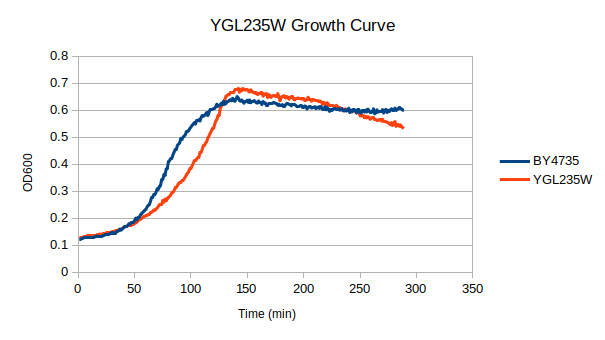
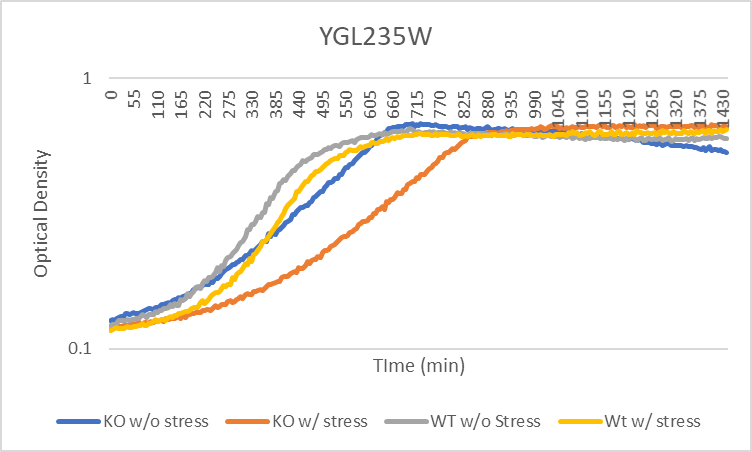
Growth Curve
In a BY4735 background, knocking out YGL235W seems to have a moderate effect on growth rate in log-phase. In this assay, the BY4735 strain's doubling time was 124 minutes, while the YGL235W knock-out strain's doubling time was 209 minutes.
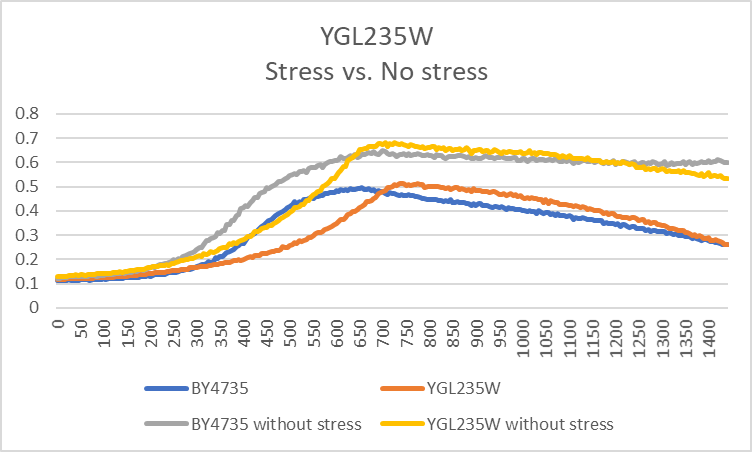
G-418 Stress
In the BY4735 background, knocking out YGL235W seems to have a little effect on the growth rate. In the knock-out experiment, the BY4735 strain's doubling time was 149 minutes, whereas the YGL235W knock-out strain's doubling time was 235 minutes. The calibration experiment, the BY4735 strain's doubling time was 64 minutes, whereas the YGL235W knock-out strain's doubling time was 93 minutes.
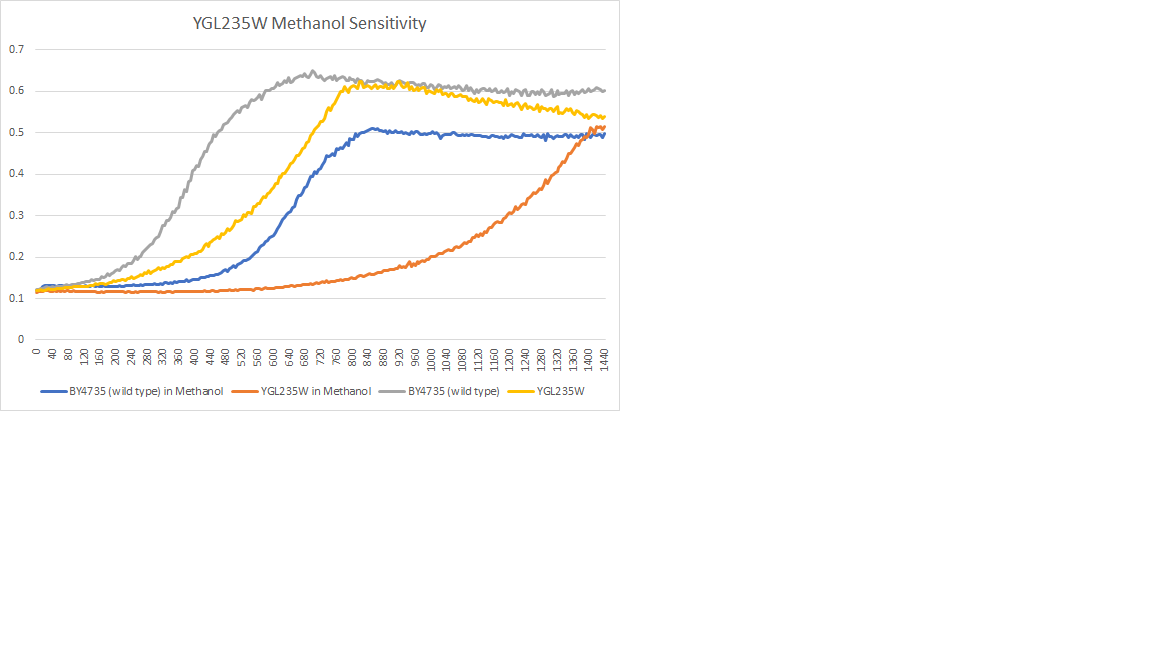
Methanol Sensitivity
The wild type had a doubling time of 160 min, the YGL235w had a doubling time of 160 min. The strain was sensitive to the methanol.
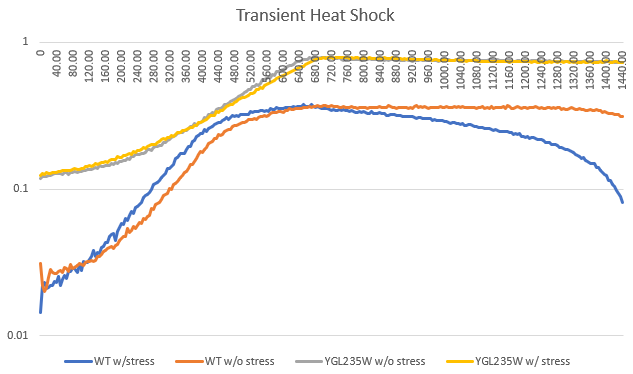
Heat Shock
Heat shock had no change on the doubling time, but the starting amount of yeast cells less. The heat seemed to kill off the cells, the longer they were in the hot water bath.
Stressing with Hydroxyurea
Hydroxyurea Protocol -The wild type strain had only a small difference in doubling time when stressed with Hydroxyurea;YGL235W had about a 20 minute difference in doubling time when stressed with hydroxyurea, indicating that this gene most likely does not play a vital role in DNA replication.
Fermentation
Fermentation protocol link [2] - There appears to be no major difference in fermentation rate. The lower final ethanol percentage could be a result of evaporation. If this experiment were to be carried out again then it should be done in an air tight container.
Caffeine Sensitivity
 Caffeine Protocol UW-Stout/Caffeine
Compared to the wild type yeast strain this gene was more sensitive to caffeine.
Caffeine Protocol UW-Stout/Caffeine
Compared to the wild type yeast strain this gene was more sensitive to caffeine.
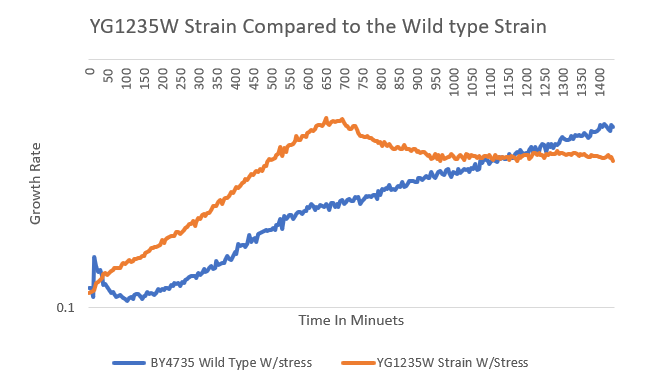
Stressing with pH
pH stress protocol link [3]
The yeast strain YGL235W had somewhat of a significant difference compared to the wild type even though the wild type keeps growing but not as fast as the yeast strain YG1235W does and the strain does plato's at the end and continues to plummets and the wild type keeps slowly climbing at a steady rate of growth and time. So, Therefore we have concluded that the data signifies that the test condition was inconclusive to the experiment.
Cold Shock Sensitivity
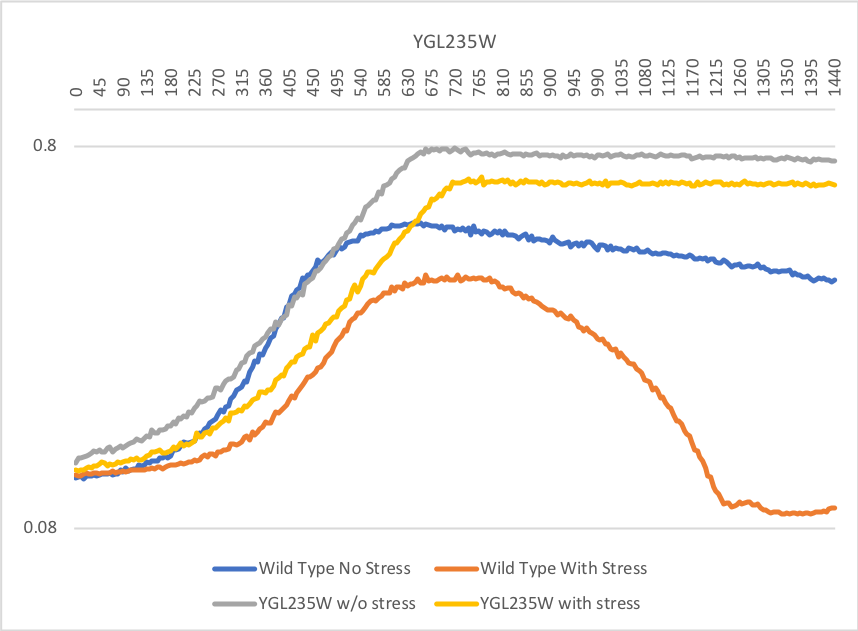 Cold Shock Protocol [[4]]
Cold Shock Protocol [[4]]
References
See Help:References on how to add references
- ↑ Chamilos G, et al. (2008) Genomewide Screening for Genes Associated with Gliotoxin Resistance and Sensitivity in Saccharomyces cerevisiae. Antimicrob Agents Chemother 52(4):1325-9 SGD PMID 18212113
- ↑ 2.0 2.1 Ubersax JA, et al. (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425(6960):859-64
SGD PMID 14574415 Cite error: Invalid
<ref>tag; name "S000074306" defined multiple times with different content - ↑ submitted by Jeff Ubersax on 2004-01-27
See Help:Categories on how to add the wiki page for this gene to a Category