Difference between revisions of "YCL002C"
SGDwikiBot (talk | contribs) (Automated import of articles) |
(→Community Commentary) |
||
| (46 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- PLEASE DO NOT EDIT HERE. USE THE SECTION EDIT LINKS ON THE RIGHT MARGIN--> | <!-- PLEASE DO NOT EDIT HERE. USE THE SECTION EDIT LINKS ON THE RIGHT MARGIN--> | ||
{{PageTop}} | {{PageTop}} | ||
| − | |||
{|{{Prettytable}} align = 'right' width = '200px' | {|{{Prettytable}} align = 'right' width = '200px' | ||
|- | |- | ||
| − | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http:// | + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http://www.yeastgenome.org/cgi-bin/locus.pl?dbid=S000000508 YCL002C] |
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | ||
| Line 13: | Line 12: | ||
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | ||
| − | |nowrap| Chr III: | + | |nowrap| Chr III:111675..110808 |
| + | |- | ||
| + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Primary SGDID''' || S000000508 | ||
|} | |} | ||
<br> | <br> | ||
| − | '''Description of | + | '''Description of YCL002C:''' Putative protein of unknown function; YCL002C is not an essential gene<ref name='S000071347'>Giaever G, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418(6896):387-91 |
{{SGDpaper|S000071347}} PMID 12140549</ref> | {{SGDpaper|S000071347}} PMID 12140549</ref> | ||
<br> | <br> | ||
| Line 23: | Line 24: | ||
<br> | <br> | ||
<br> | <br> | ||
| − | |||
__TOC__ | __TOC__ | ||
==Community Commentary== | ==Community Commentary== | ||
| Line 29: | Line 29: | ||
| − | < | + | ===[[UW-Stout/Sensitivity To Nitrogen Starvation]]=== |
| + | [[File: YCLO02C_growth plate.JPG|thumb|Growth of YCLO02C on synthetic defined media (SDC).]] | ||
| + | [[File: YCL002C starvation plate1.JPG|thumb|Growth of YCLO02C on omission media that lacks nitrogen.]] | ||
| + | Knocking out the YCLO02C seems to have no effect on growth after incubating for 5 days on the Nitrogen omitted media. | ||
| + | |||
| + | ===[[UW-Stout/Ultraviolet Light|Ultraviolet Sensitivity]]=== | ||
| + | |||
| + | [[File:YCL002C uv light.jpg.jpg|300 px]] | ||
| + | [[File:BY4735(wild) uv light.jpg|300 px]] | ||
| + | |||
| + | YCL002C is more sensitive than BY4735(wild) given that there is less colonizes present when under the same stress. | ||
| + | |||
| + | |||
| + | |||
| + | {{UW-Stout}} | ||
| + | |||
| + | ===[[UW-Stout/Growth_Curve|Growth Rate in YPD]]=== | ||
| + | |||
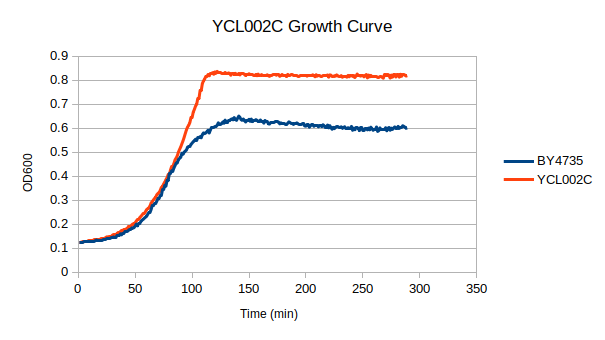
| + | [[File:ycl002c_growth.png]] | ||
| + | |||
| + | In a BY4735 background, knocking out YCL002C seems to have no effect on growth rate in log-phase in YPD. In this assay, the BY4735 strain's doubling time was 124 minutes, while the YCL002C knock-out strain's doubling time was 134 minutes. However, the knock-out strain did reach a substantially higher OD600 before saturating. | ||
| + | |||
| + | ===[[UW-Stout/G-418 | G-418 Stress]]=== | ||
| + | |||
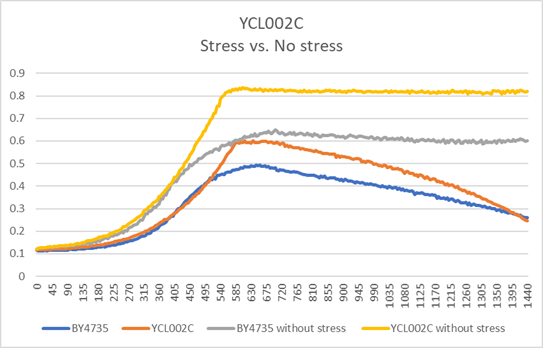
| + | [[File:image017.png]] | ||
| + | |||
| + | In the BY4735 background, knocking out YCL002C seems to have a little effect on the growth rate by slowing it down. In the knock-out experiment, the BY4735 strain's doubling time was 149 minutes, whereas the YCL002C knock-out strain's doubling time was 172 minutes. The calibration experiment, the BY4735 strain's doubling time was 64 minutes, whereas the YCL002C knock-out strain's doubling time was 66 minutes. | ||
| + | |||
| + | ===Methanol Sensitivity=== | ||
| + | |||
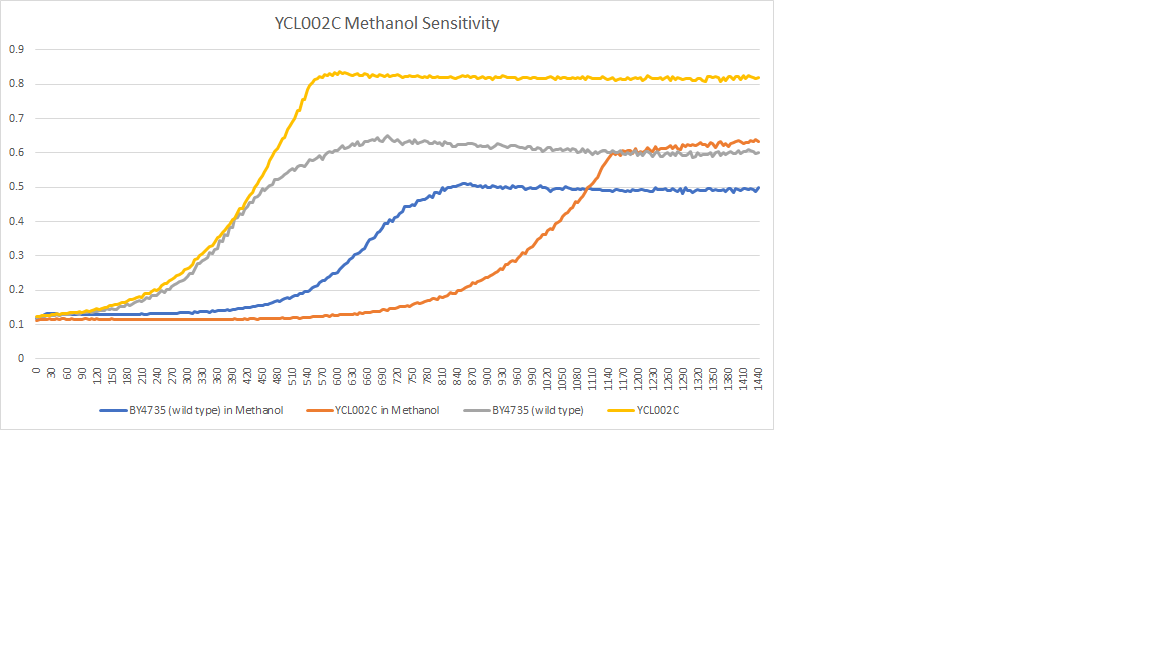
| + | The wild type had a doubling time of 175 minutes, the YCL002 had a doubling time of 315 minutes. The addition of methanol sped up our reaction and caused the yeast cells to multiply faster. | ||
| + | |||
| + | [https://wiki.yeastgenome.org/index.php/UW-Stout/Methanol] | ||
| + | |||
| + | [[File:YCL002C_Methanol.png]] | ||
| + | |||
| + | ===Stressing with Hydroxyurea=== | ||
| + | [[UW-Stout/Hydroxyurea|Hydroxyurea Protocol]] | ||
| + | -The wild type strain had only a small difference in doubling time when stressed with Hydroxyurea;YCL002C also portrayed a small difference in doubling time, indicating that this gene most likely does not play a role in DNA replication. | ||
| + | [[file:HU_YCL002C.png|left]] | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | ===Stressing with pH=== | ||
| + | pH stress protocol link [https://wiki.yeastgenome.org/index.php/UW-Stout/pH] | ||
| + | |||
| + | |||
| + | - The wild type strain had a significant difference in doubling time when my partner and I stressed it out with a varying levels of acidic pH's and It displayed that when we tested it on the final lab experiment of a pH of 4 that it was highly sensitive to that particular pH and In some of the test conditions that it displayed that they have died or are very very slowly growing. But the yeast gene strain YCL002C we came to the conclusion that it was a lot more resilient to the acidic pH of 4 than the wild type strain. | ||
| + | |||
| + | |||
| + | [[File:Examples.PNG]] | ||
| + | |||
| + | ===Transient Heat Shock=== | ||
| + | |||
| + | |||
| + | [[File:YCL002C Heat Shock.PNG]] | ||
| + | |||
| + | Heat shock had no change on the doubling time, but the starting amount of yeast cells less. The heat seemed to kill off the cells, the longer they were in the hot water bath. | ||
| + | |||
| + | ===Fermentation=== | ||
| + | Fermentation protocol link [https://wiki.yeastgenome.org/index.php/UW-Stout/Fermentation] | ||
| + | - There appears to be no major difference in fermentation rate. The lower final ethanol percentage could be a result of evaporation. If this experiment were to be carried out again then it should be done in an air tight container. | ||
| + | |||
| + | [[File:Fer YGL002C.PNG]] | ||
| + | |||
| + | <!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | ||
| + | <!-- | ||
| + | Specifically higher expression in carbon limited chemostat cultures versus carbon excess. | ||
| + | <ref>Boer VM, et al. (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. | ||
| + | J Biol Chem 278(5):3265-74</ref> | ||
| + | --> | ||
| + | |||
| + | ===Caffeine Sensitivity=== | ||
| + | |||
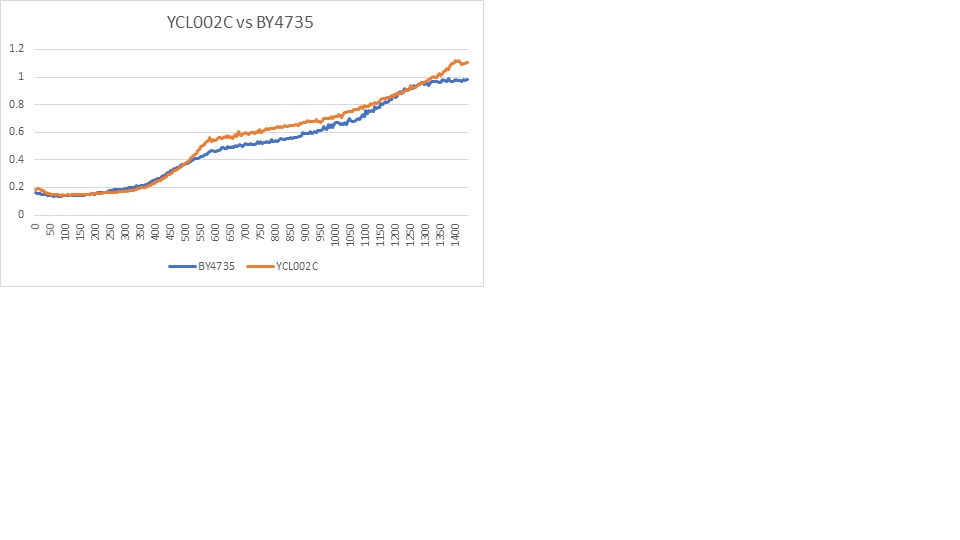
| + | [[File:YCL002C_vs_BY4735.jpg|YCL002C VS BY4735]] | ||
| + | |||
| + | Caffeine experiment protocol [[UW-Stout/Caffeine]] | ||
| + | |||
| + | It appears the caffeine concentration had a very similar effect on this gene and the wild type yeast cells. | ||
| + | |||
| + | ===Cold Shock Sensitivity=== | ||
| + | [[File:YCL002C.png]] | ||
| + | |||
| + | |||
| + | Protocol: [https://wiki.yeastgenome.org/index.php/UW-Stout/Cold_Shock Cold Shock] | ||
| + | |||
| + | |||
| + | |||
| + | YCL002C is neither more or less sensitive to the cold than the wild type strain. This is shown because the doubling times change the same amount before and after the cold shock in both the wild type strain and YCL002C. | ||
| + | |||
==References== | ==References== | ||
<!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | <!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | ||
{{RefHelp}} | {{RefHelp}} | ||
| − | |||
Latest revision as of 18:37, 9 May 2019
Share your knowledge...Edit this entry!
| Systematic name | YCL002C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr III:111675..110808 |
| Primary SGDID | S000000508 |
Description of YCL002C: Putative protein of unknown function; YCL002C is not an essential gene[1]
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
UW-Stout/Sensitivity To Nitrogen Starvation
Knocking out the YCLO02C seems to have no effect on growth after incubating for 5 days on the Nitrogen omitted media.
Ultraviolet Sensitivity
YCL002C is more sensitive than BY4735(wild) given that there is less colonizes present when under the same stress.
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
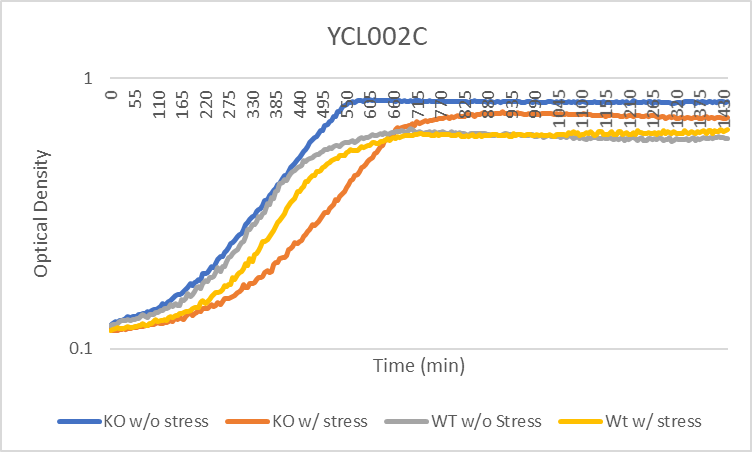
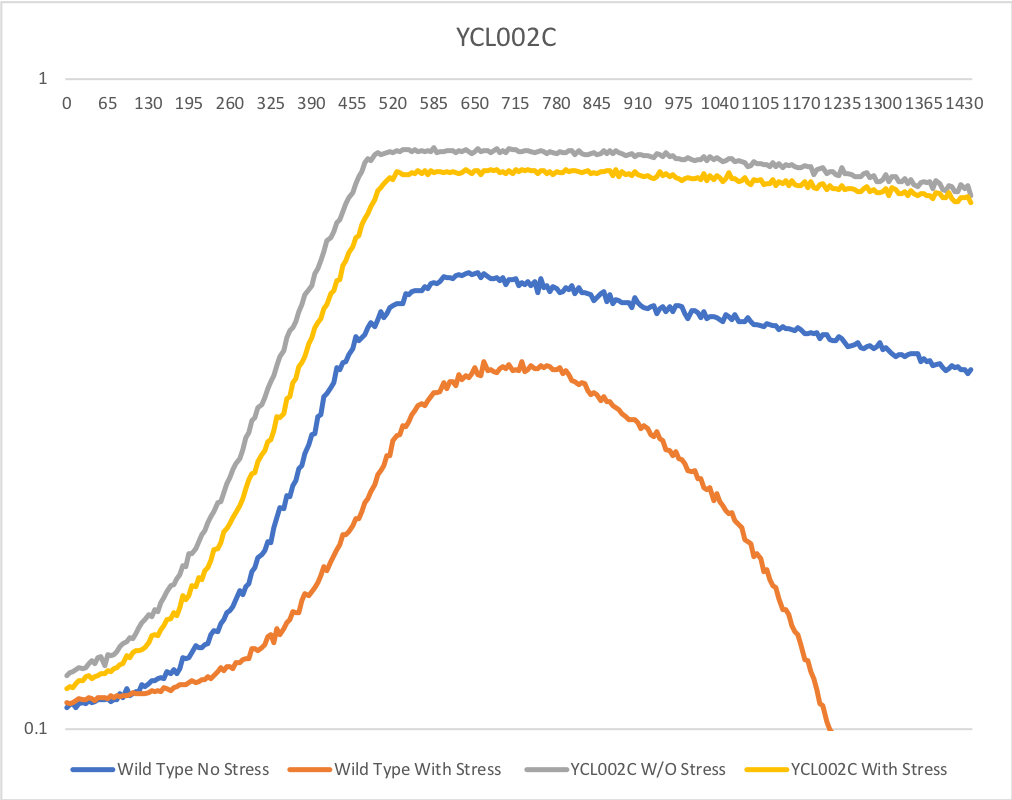
Growth Rate in YPD
In a BY4735 background, knocking out YCL002C seems to have no effect on growth rate in log-phase in YPD. In this assay, the BY4735 strain's doubling time was 124 minutes, while the YCL002C knock-out strain's doubling time was 134 minutes. However, the knock-out strain did reach a substantially higher OD600 before saturating.
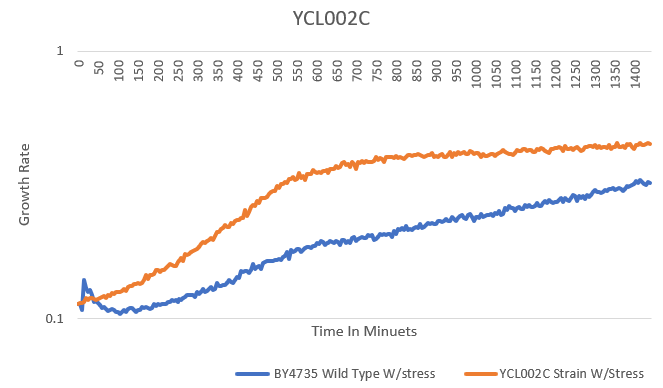
G-418 Stress
In the BY4735 background, knocking out YCL002C seems to have a little effect on the growth rate by slowing it down. In the knock-out experiment, the BY4735 strain's doubling time was 149 minutes, whereas the YCL002C knock-out strain's doubling time was 172 minutes. The calibration experiment, the BY4735 strain's doubling time was 64 minutes, whereas the YCL002C knock-out strain's doubling time was 66 minutes.
Methanol Sensitivity
The wild type had a doubling time of 175 minutes, the YCL002 had a doubling time of 315 minutes. The addition of methanol sped up our reaction and caused the yeast cells to multiply faster.
Stressing with Hydroxyurea
Hydroxyurea Protocol -The wild type strain had only a small difference in doubling time when stressed with Hydroxyurea;YCL002C also portrayed a small difference in doubling time, indicating that this gene most likely does not play a role in DNA replication.
Stressing with pH
pH stress protocol link [2]
- The wild type strain had a significant difference in doubling time when my partner and I stressed it out with a varying levels of acidic pH's and It displayed that when we tested it on the final lab experiment of a pH of 4 that it was highly sensitive to that particular pH and In some of the test conditions that it displayed that they have died or are very very slowly growing. But the yeast gene strain YCL002C we came to the conclusion that it was a lot more resilient to the acidic pH of 4 than the wild type strain.
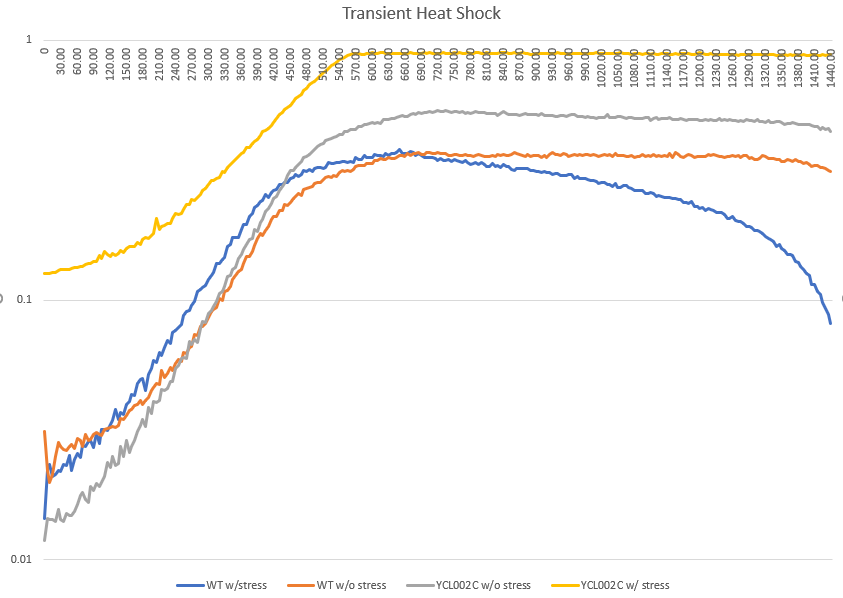
Transient Heat Shock
Heat shock had no change on the doubling time, but the starting amount of yeast cells less. The heat seemed to kill off the cells, the longer they were in the hot water bath.
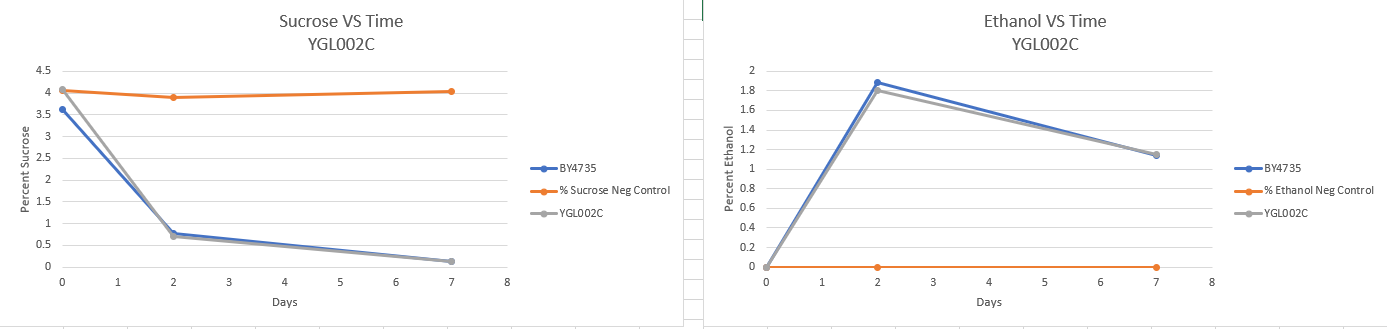
Fermentation
Fermentation protocol link [3] - There appears to be no major difference in fermentation rate. The lower final ethanol percentage could be a result of evaporation. If this experiment were to be carried out again then it should be done in an air tight container.
Caffeine Sensitivity
Caffeine experiment protocol UW-Stout/Caffeine
It appears the caffeine concentration had a very similar effect on this gene and the wild type yeast cells.
Cold Shock Sensitivity
Protocol: Cold Shock
YCL002C is neither more or less sensitive to the cold than the wild type strain. This is shown because the doubling times change the same amount before and after the cold shock in both the wild type strain and YCL002C.
References
See Help:References on how to add references
See Help:Categories on how to add the wiki page for this gene to a Category