Difference between revisions of "YDR307W"
(→Community Commentary) |
(→UW-Stout/Hydrogen Peroxide SP22) |
||
| Line 161: | Line 161: | ||
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to hydrogen peroxide. | As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to hydrogen peroxide. | ||
| + | |||
| + | ====Results==== | ||
| + | |||
| + | BY4735 and YDR307W after being exposed to 0.065% dilution of hydrogen peroxide solution. | ||
| + | |||
| + | [[File:YDR307Wperosm.png]] | ||
| + | |||
| + | ====Interpretation==== | ||
==References== | ==References== | ||
Revision as of 17:46, 4 May 2022
Share your knowledge...Edit this entry! <protect>
| Systematic name | YDR307W |
| Gene name | PMT7 |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr IV:1075865..1077853 |
| Primary SGDID | S000002715 |
Description of YDR307W: Putative protein mannosyltransferase similar to Pmt1p; has a potential role in protein O-glycosylation[1]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
UW Stout/D2O SP22
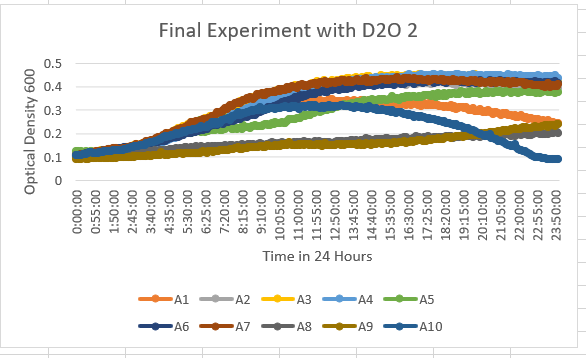
The YDR307W Knock Out Yeast Strain is A5 and is fairly unaffected by the 35% dilution of D2O, however, has a slight sensitivity to the dilution. With the maximum value ranging along the lines of 0.4-0.5 OD 600.
UW Stout/Sucrose Fermentation SP22
| Gene | Glucose | Fructose | Ethanol |
| Standard solution | 2.0000 | 0.2000 | 2.0000 |
| YDl109C | 0.3800 | 0.3933 | 0.3430 |
| YGL140C | 0.2212 | 0.2685 | 0.1867 |
| YOR111W | 0.3332 | 0.3598 | 0.1343 |
| YHL029C | 0.3870 | 0.2368 | 0.1151 |
| YDR307W | 0.4366 | 0.2487 | 0.0606 |
| YNL058C | 0.2710 | 0.3056 | 0.1577 |
| YCL049C | 0.4078 | 0.3052 | 0.1969 |
| YGR079W | 0.4042 | 0.1589 | 0.0080 |
| YBL113C | 0.3498 | 0.2012 | 0.1434 |
| BY4735 | 0.3171 | 0.3084 | 0.3541 |
Interpretation
This gene produced 17.11% of the amount of ethanol that the wild type produced. It is possible that this gene plays a role in fermentation because of such a low amount of ethanol production.
<protect>
UW-Stout/UV Light SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested under a UV Light using this protocol.
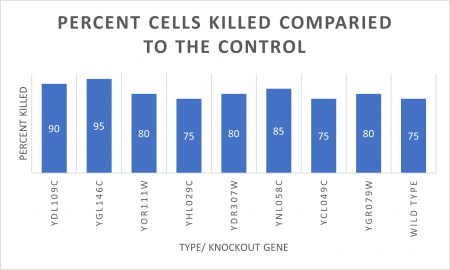
RESULTS
INTERPERTATION
In the graph and photos above, exposing this gene to 600 seconds of 400 Watt UV Light killed approximately 80% of yeast cell cultures, compared to its control counterpart, which was the same gene and amount of cells, just was not exposed to UV Light.
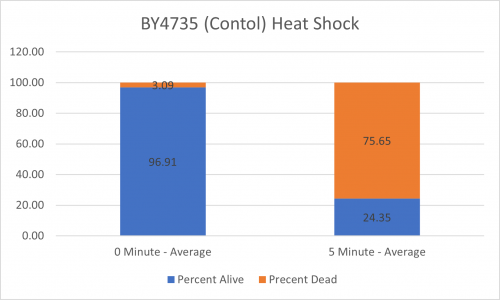
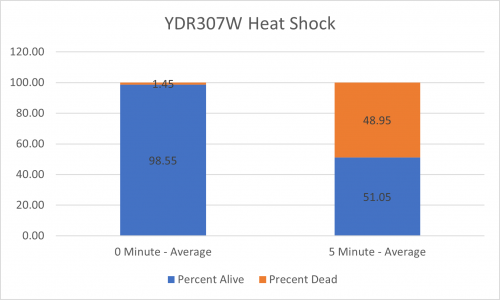
UW-Stout/Heat Shock SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to heat shock.
Results
Interpretation
From the data gathered, there was a medium positive effect to knocking out this gene when it came to the yeast's ability to hold up to heat shock. The modified yeast cells were 210% more resistant to heat shock than the control.
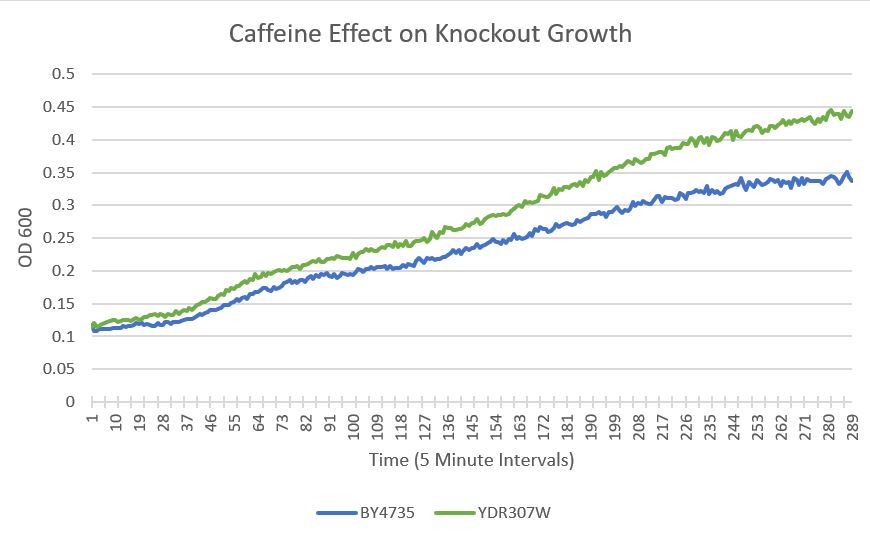
UW-Stout/Caffeine SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to 4mM of caffeine.
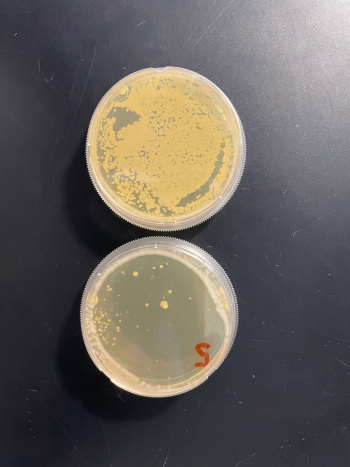
Results
- BY4735(wild type yeast) and YDR307W(knockout yeast gene) growth after being subjected to 4mM caffeine solution.
Interpretation
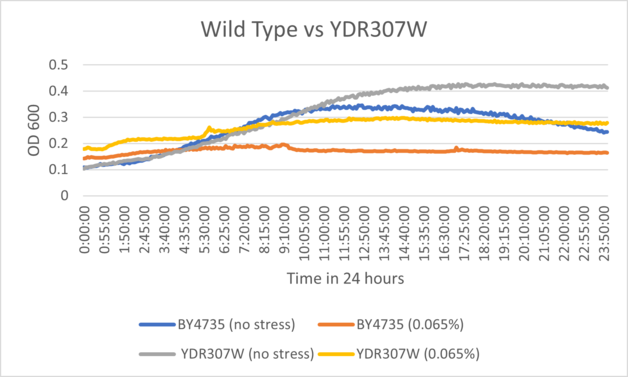
UW-Stout/Hydrogen Peroxide SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to hydrogen peroxide.
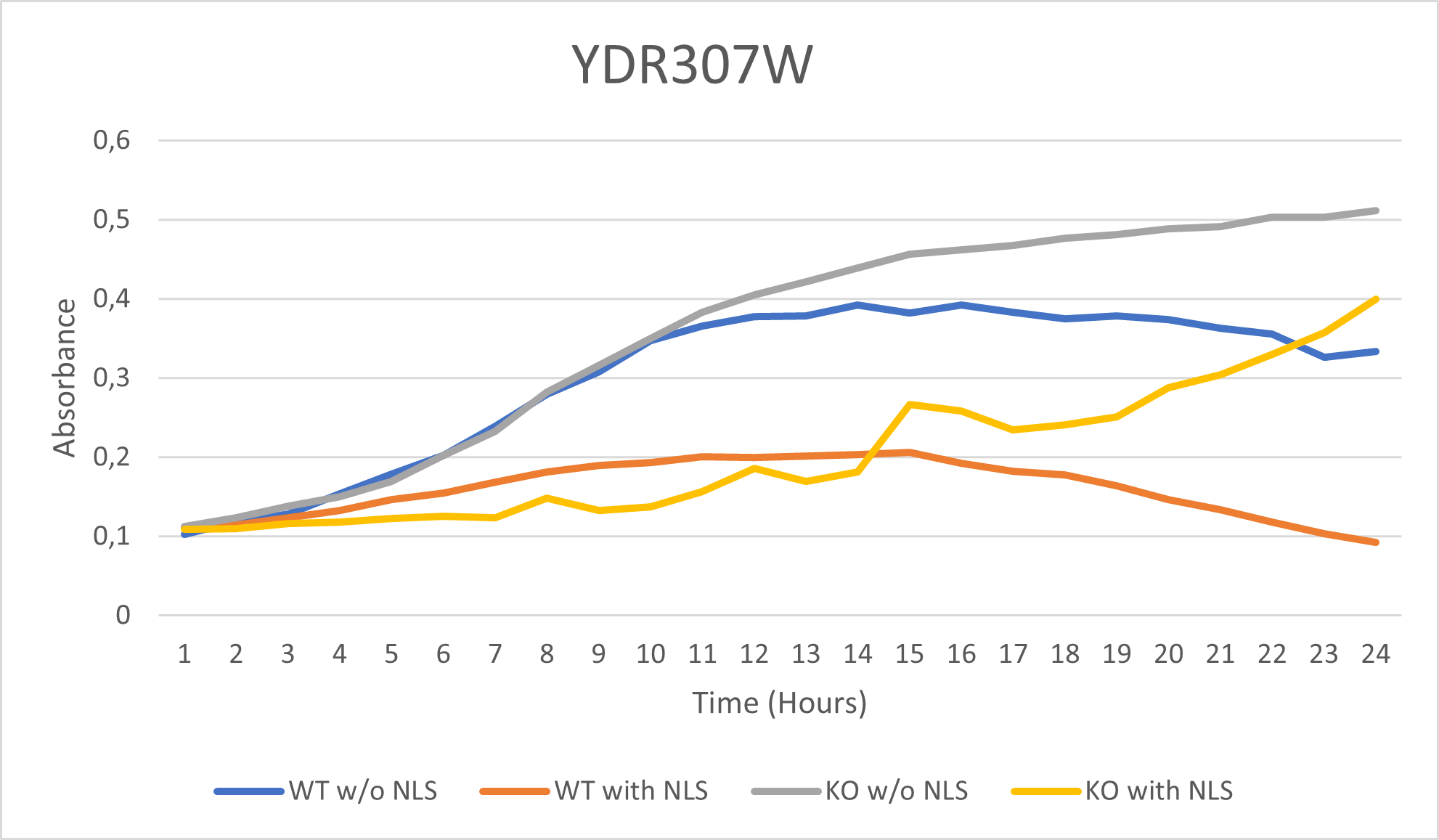
Results
BY4735 and YDR307W after being exposed to 0.065% dilution of hydrogen peroxide solution.
Interpretation
References
See Help:References on how to add references
See Help:Categories on how to add the wiki page for this gene to a Category </protect>