Difference between revisions of "YGL138C"
SGDwikiBot (talk | contribs) (Automated import of articles) |
(→Doubling Time at 0.25 concentration) |
||
| (54 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
{|{{Prettytable}} align = 'right' width = '200px' | {|{{Prettytable}} align = 'right' width = '200px' | ||
|- | |- | ||
| − | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http:// | + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http://www.yeastgenome.org/cgi-bin/locus.pl?dbid=S000003106 YGL138C] |
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | ||
| − | |nowrap| Chr VII: | + | |nowrap| Chr VII:249531..248494 |
| + | |- | ||
| + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Primary SGDID''' || S000003106 | ||
|} | |} | ||
<br> | <br> | ||
| − | '''Description of | + | '''Description of YGL138C:''' Putative protein of unknown function; has no significant sequence similarity to any known protein<ref name='S000043179'>Victoria Escribano M and Mazon MJ (2000) Disruption of six novel ORFs from Saccharomyces cerevisiae chromosome VII and phenotypic analysis of the deletants. Yeast 16(7):621-30 |
| + | {{SGDpaper|S000043179}} PMID 10806424</ref> | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 27: | Line 30: | ||
{{CommentaryHelp}} | {{CommentaryHelp}} | ||
| + | {{UW-Stout}} | ||
| + | |||
| + | ===[[UW-Stout/Growth_Curve_SP21|Growth Curve]]=== | ||
| + | |||
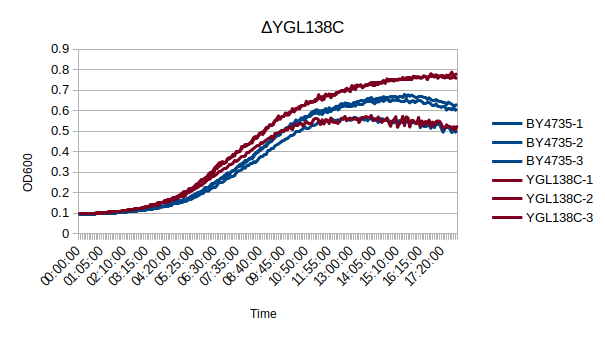
| + | [[Image:YGL138C-water.png]] | ||
| + | |||
| + | In a BY4735 background, knocking out YGL138C does not seem to have much effect on the strain's growth rate. In this assay, the BY4735 strain's doubling time was 162 minutes, while the YGL138C knock-out strain's doubling time was 140 minutes. (These doubling times are the means of three experiments.) | ||
| + | |||
| + | ===[[UW-Stout/Hydroxychloroquine_SP21|Hydroxychloroquine]]=== | ||
| + | |||
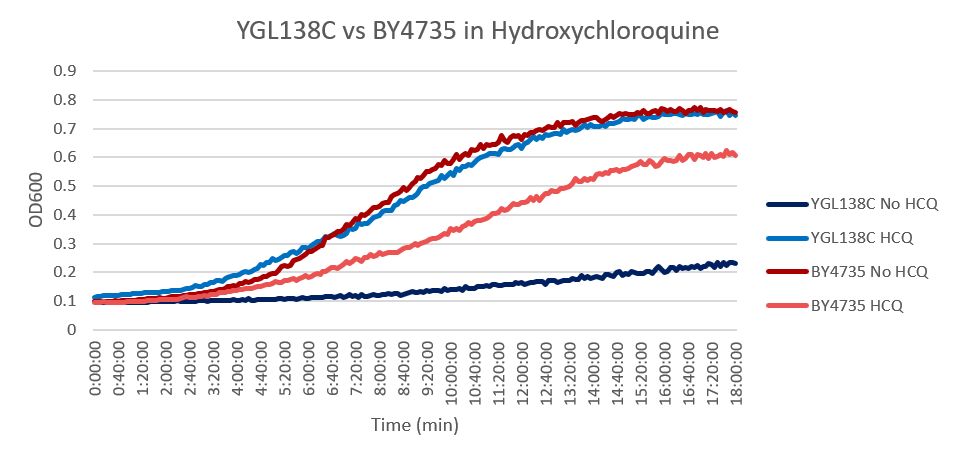
| + | [[Image:YGL138C-BY4735.png]] | ||
| + | |||
| + | HCQ refers to hydroxychloroquine | ||
| + | |||
| + | Under normal conditions, the BY4735 doubling time is 144 minutes, while that of YGL138C is 743 minutes. Thus, YGL138C doubles much slower than the wild strain. When grown in an environment with hydroxychloroquine, YGL138C doubled in 224 minutes, while BY4735 doubled in 276 minutes. Interestingly, YGL138C doubling time decreased dramatically, which means that it grew faster in the presence of hydroxychloroquine. The lag phase of YGL138C also decreased meaning that rapid growth started earlier than that of the YGL138C under normal conditions. The BY4735 strain was inhibited by hydroxychloroquine. (These doubling times and curves are the means of three experiments.) | ||
| + | |||
| + | ===[[UW-Stout/PMSF SP21|Phenylmethylsulphonyl Fluoride]]=== | ||
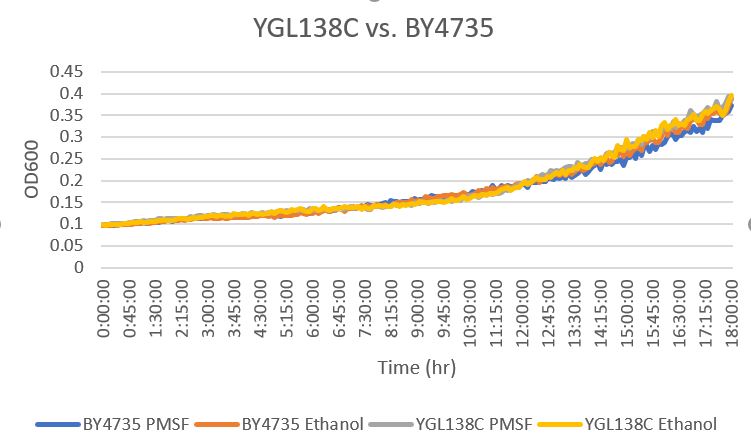
| + | [[File:YGL138C vs Wild type.jpg]] | ||
| + | The YGL138C yeast gene strain was determined to be barely different in growth to the BY4735 strain. The YGL138C strain had a doubling time of 373 minutes while the BY4735 strain has a doubling time of 382 minutes. This means that the modification had little to no effect on the way that the cell interacts with PMSF. The doubling time of the BY4735 with ethanol had a doubling time of 442 minutes, while the YGL138C with ethanol had a doubling time of 351 minutes. | ||
| + | ===[[UW-Stout/Sodium Hydroxide SP21|Sodium Hydroxide]]== | ||
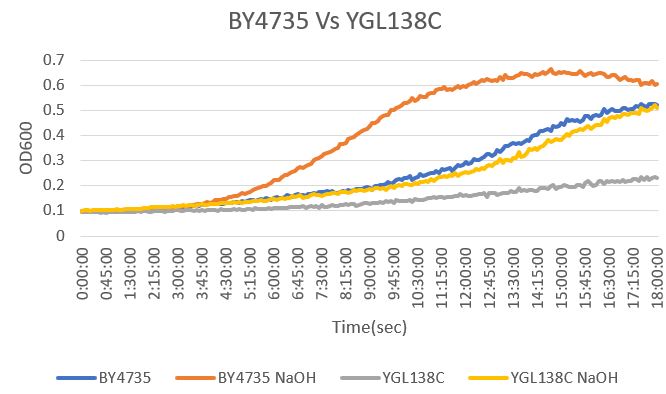
| + | [[File:YGL vs Wt.jpg]] | ||
| + | The YGL138C yeast strain had a significant difference from the BY4735 that were both exposed to NaOH. The YGL138C strain had a doubling time of 363 minutes while the BY4735 strain had a doubling time of 545 minutes. That means that the NaOH affected the growth of the YGL138C significantly. The doubling time for BY4735 is 809 minutes and for YGL138C is 785 minutes. | ||
<!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | <!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | ||
<!-- | <!-- | ||
| Line 36: | Line 61: | ||
J Biol Chem 278(5):3265-74</ref> | J Biol Chem 278(5):3265-74</ref> | ||
--> | --> | ||
| + | ---- | ||
| + | |||
| + | ===[[UW-Stout/Salt_Concentration_SP21|Salt Concentration (NaCl)]]=== | ||
| + | |||
| + | [[Image:NaCl-BY4735vYGL138C-Graph.JPG]] | ||
| + | |||
| + | |||
| + | 0mM NaCl conc. BY4735 strain's doubling time: 169 minutes | ||
| + | |||
| + | 0mM NaCl conc. YGL138C strain's doubling time: 139 minutes | ||
| + | |||
| + | 750mM NaCl conc. BY4735 strain's doubling time: 294 minutes | ||
| + | |||
| + | 750mM NaCl conc. YGL138C strain's doubling time: 205 minutes | ||
| + | |||
| + | |||
| + | The graph above shows the growth rate for the previously listed strains and the relative level of NaCl concentration. Knocking out the gene positively influenced the growth rate. Comparing the YGL138C doubling times in 0mM and 750mM NaCl, there is a moderate effect on the growth rate. This effect negatively impacts the knockout strain's (YGL138C) growth rate. | ||
| + | ---- | ||
| + | |||
| + | ===[[UW-Stout/Caffeine_SP21|Caffeine Group 1]]=== | ||
| + | [[File:gnevswild6.jpg|center|thumb|500px]] | ||
| + | |||
| + | When adding caffeine to the YGL138C strain, the effect it had on the growth rate was significant. The doubling times are as follows: | ||
| + | |||
| + | Doubling time of YGL138C with caffeine: 309 minutes | ||
| + | |||
| + | Doubling time of YGL138C without caffeine: 140 minutes | ||
| + | |||
| + | Doubling time of BY4735 with caffeine: 138 minutes | ||
| + | |||
| + | Doubling time of BY4735 without caffeine: 187 minutes | ||
| + | |||
| + | When adding caffeine to this strain knockout, the growth rate slowed down a great deal. | ||
| + | |||
| + | ===[[UW-Stout/Co Culture SP21|Competative Co-culture]]=== | ||
| + | |||
| + | [[File:YGL138CCompetetiveCoCulture.jpg]] | ||
| + | |||
| + | These are the results of a competitive co-culture protocol. The knockout strain was grown in a culture to test fitness against a green, fluorescent wild-type strain. The percentage of analyzed cells in the culture measured was those displaying green fluorescence. In theory, this should mean if a knockout has reduced the fitness of a strain of yeast, the fluorescent wild-type strain would have a higher percentage than the knockout strain. If the knockout does not decrease fitness, they would be roughly equal. This may not be so in results. Sources of error may include contamination and human error. | ||
| + | |||
| + | The results here are not expected in theory. The YGL138C knockout strain had a significant disadvantage and the fluorescent wild-type strain dominated the given result. Test 1 must have been contaminated with a faster growing organism than yeast that did not display a fluorescent behavior like the wild-type fluorescent yeast used. However, Strain 2 appears to follow an expected result and suggests the knockout strain had a significant disadvantage imposed to its fitness, given the wild-type strain outgrew it. | ||
| + | |||
| + | ===Ammonium Sulfate=== | ||
| + | |||
| + | [[File:TNKO6.jpg|center|thumb|500px]] | ||
| + | |||
| + | YGL138C gene shows an interesting result. While the wild strain from 5g/ml of ammonium sulfate to none shows a 17.8% slower rate, it shows a 35.5% slower rate. For further analysis 0.5g/ml to no ammonium sulfate was compared too. Wild showed 1% slower rate and YGL138C showed 38% slowed rate. Based on the differences of these two treatments in the wild to the YGL138C KO strain it could be said that there was experimental error. The differences in percent are 16.8% in the wild and roughly 3% in the KO strain. The doubling time of the negative control is like the wild as well. It could be that errors in volume transfers occurred causing increased growth in the other treatments. It could also be that this strain with the knocked out YGL138C gene is in fact more sensitive to nitrogen starvation. | ||
<protect> | <protect> | ||
| + | ---- | ||
| + | ===[[UW-Stout/UV Light SP21|UV Light]]=== | ||
| + | [[Image:D61CE5D0-5F3F-427C-B899-9942215B07DB.jpeg|center|thumb|400px|Knock-out Strain 6]] | ||
| + | '''Results''': | ||
| + | *Experiment 1:0sec=437 colonies, 600sec=81 colonies. | ||
| + | *Experiment 2:0sec=135 colonies, 600sec=28 colonies. | ||
| + | '''Interpretation''': The two 0sec plates are on top and the 600sec plates are on the bottom. The 0sec yeast plate from experiment 2 had a lot of contamination along with the yeast while the 600sec plate from experiment 2 had very little. This yeast strain was about 80% for both plates which is more significant than some of the other strains however I'm not sure I'd say this is significant. It's possible that the number of yeast colonies was miscounted on the 0sec experiment 2 plate because of how small many of colonies are and how much contamination there is. Though it may be possible the gene played a role in UV resistance, it's hard to say with only one run of the experiment being good. | ||
| + | |||
| + | |||
| + | ---- | ||
| + | ===[[UW-Stout/Calcofluor White SP21|Calcofluor White]]=== | ||
| + | [[Image:YGL138C CW.jpg|YGL138 CW]] | ||
| + | |||
| + | From this data we can determine that there was moderate growth inhibition on this strain of yeast. The modified yeast had a doubling rate that was about twenty minutes slower than the wild type on average, and when compared to the effect on other yeast strains this is a moderate effect on the inhibition of growth. | ||
| + | ---- | ||
| + | |||
| + | ===[[UW-Stout/Heat Shock SP21|Heat Shock]]=== | ||
| + | [[File:thumbnail_IMG_3522.jpg|center|thumb|500px|YGL138C yeast strain in pitri dishes.]] | ||
| + | |||
| + | The yeast strain YGL138C was impacted significantly by the heat shocking of the thermal cycler. As seen in the image, the controlled plate grew a lot of small DNA colonies all throughout the majority of pitri dish. It looks like a very thick sheet of cells. When compared to the two trial heat shocking plates, there are a lot less cells being grown. The trial cells are slightly larger than those of the controlled, but not by much. The quantity of the cells is much smaller, and the quality (size) of the cells is not that much bigger than what they're being compared to. This gene was heat sensitive, but not as much as the YLR426W yeast strain. | ||
| + | ---- | ||
| + | ===[[UW-Stout/Caffeine 2 SP21|Caffeine Group 2]]=== | ||
| + | [[File:YGL138C graph.png|700px]] | ||
| + | ====Doubling Time at 0.25 concentration==== | ||
| + | YGL138C Td = (217-1) x ln(2)/ln(0.2/0.1) = 216 minutes; YGL138C Td= (153-33) x ln(2)/ln(.7/.1)= 42 minutes; Wild Type Td = (146-66) x ln(2)/ln(.6/.2) = 88 minutes These doubling times show that the 0.25 mM concentration in YGL138C slowed down the rate of growth. The caffeine was able to slow down metabolic activity. Caffeine may effect the function of the Rad52 protein that aids in DNA double-stranded breaks and maintaining genome stability. The caffeine may also aid in the process of the cell protecting itself from reacting oxygen species. Since the concentration is small in volume, apoptosis was not induced for cell function. | ||
| + | |||
| + | ===[[UW-Stout/Ethanol Sensitivity| Ethanol Sensitivity]]=== | ||
| + | [[File:YGL138C Ethanol.png]] | ||
| + | |||
| + | The YGL138C yeast strain was determined to be barely different in growth to the BY4735 strain. The YGL138C strain had a doubling time of 191 minutes while the BY4735 strain has a doubling time of 182 minutes. This means that the modification had little to no effect on the way that the cell interacts with ethanol. The doubling time of the BY4735 with ethanol had a doubling time of 218 minutes, while the YGL138C with ethanol had a doubling time of 212 minutes. | ||
| + | |||
| + | [[https://wiki.yeastgenome.org/index.php/UW-Stout/Ethanol_SP21]] | ||
| + | |||
| + | |||
| + | |||
| + | == Hyrdoxyurea == | ||
| + | [[File:YGL.png|center|1000px|]] | ||
| + | |||
| + | The YGL138X strain had an average doubling time that was very similar to the growth rate of the BY4735 strain. The stressed YGL138C strain had a doubling time of 286 minutes, while the average doubling time of the stressed BY4735 strain had an average doubling time of 278 minutes. What is similar in both unstressed samples is what is interesting, the unstressed YGL138C strain had an average doubling time of 167 minutes, while the BY4735 unstressed strain had an average doubling time of 182 minutes. This means the stressor had little effect on the rate of growth of the strain. | ||
| + | [https://wiki.yeastgenome.org/index.php/UW-Stout/Hydroxyurea_SP21] | ||
| + | |||
==References== | ==References== | ||
<!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | <!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | ||
{{RefHelp}} | {{RefHelp}} | ||
</protect> | </protect> | ||
Latest revision as of 07:07, 5 May 2021
Share your knowledge...Edit this entry! <protect>
| Systematic name | YGL138C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr VII:249531..248494 |
| Primary SGDID | S000003106 |
Description of YGL138C: Putative protein of unknown function; has no significant sequence similarity to any known protein[1]
</protect>
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
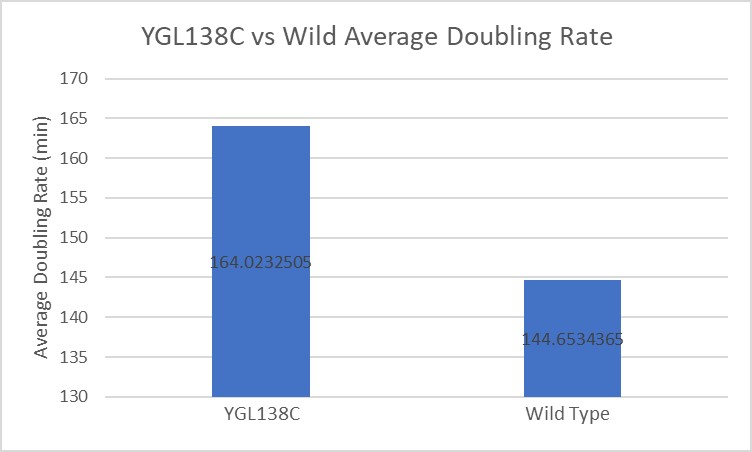
Growth Curve
In a BY4735 background, knocking out YGL138C does not seem to have much effect on the strain's growth rate. In this assay, the BY4735 strain's doubling time was 162 minutes, while the YGL138C knock-out strain's doubling time was 140 minutes. (These doubling times are the means of three experiments.)
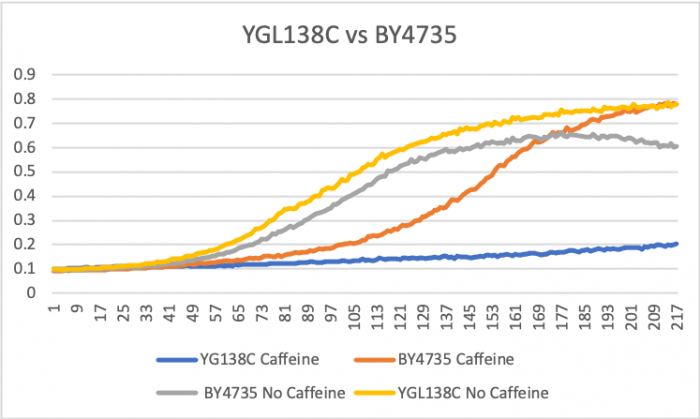
Hydroxychloroquine
HCQ refers to hydroxychloroquine
Under normal conditions, the BY4735 doubling time is 144 minutes, while that of YGL138C is 743 minutes. Thus, YGL138C doubles much slower than the wild strain. When grown in an environment with hydroxychloroquine, YGL138C doubled in 224 minutes, while BY4735 doubled in 276 minutes. Interestingly, YGL138C doubling time decreased dramatically, which means that it grew faster in the presence of hydroxychloroquine. The lag phase of YGL138C also decreased meaning that rapid growth started earlier than that of the YGL138C under normal conditions. The BY4735 strain was inhibited by hydroxychloroquine. (These doubling times and curves are the means of three experiments.)
Phenylmethylsulphonyl Fluoride
The YGL138C yeast gene strain was determined to be barely different in growth to the BY4735 strain. The YGL138C strain had a doubling time of 373 minutes while the BY4735 strain has a doubling time of 382 minutes. This means that the modification had little to no effect on the way that the cell interacts with PMSF. The doubling time of the BY4735 with ethanol had a doubling time of 442 minutes, while the YGL138C with ethanol had a doubling time of 351 minutes.
=Sodium Hydroxide
The YGL138C yeast strain had a significant difference from the BY4735 that were both exposed to NaOH. The YGL138C strain had a doubling time of 363 minutes while the BY4735 strain had a doubling time of 545 minutes. That means that the NaOH affected the growth of the YGL138C significantly. The doubling time for BY4735 is 809 minutes and for YGL138C is 785 minutes.
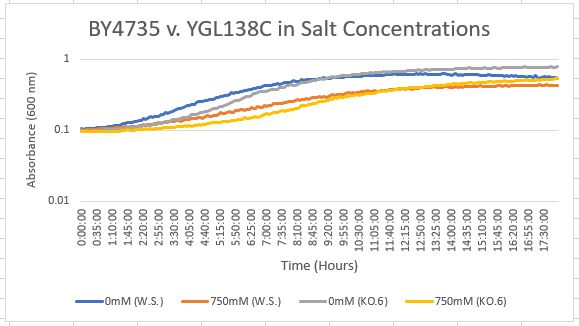
Salt Concentration (NaCl)
0mM NaCl conc. BY4735 strain's doubling time: 169 minutes
0mM NaCl conc. YGL138C strain's doubling time: 139 minutes
750mM NaCl conc. BY4735 strain's doubling time: 294 minutes
750mM NaCl conc. YGL138C strain's doubling time: 205 minutes
The graph above shows the growth rate for the previously listed strains and the relative level of NaCl concentration. Knocking out the gene positively influenced the growth rate. Comparing the YGL138C doubling times in 0mM and 750mM NaCl, there is a moderate effect on the growth rate. This effect negatively impacts the knockout strain's (YGL138C) growth rate.
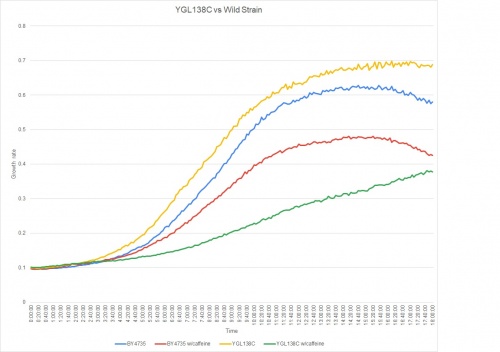
Caffeine Group 1
When adding caffeine to the YGL138C strain, the effect it had on the growth rate was significant. The doubling times are as follows:
Doubling time of YGL138C with caffeine: 309 minutes
Doubling time of YGL138C without caffeine: 140 minutes
Doubling time of BY4735 with caffeine: 138 minutes
Doubling time of BY4735 without caffeine: 187 minutes
When adding caffeine to this strain knockout, the growth rate slowed down a great deal.
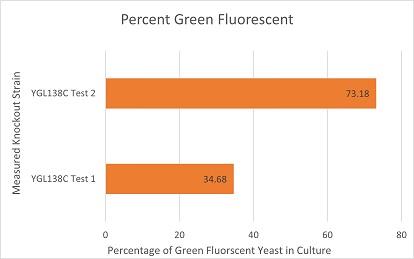
Competative Co-culture
These are the results of a competitive co-culture protocol. The knockout strain was grown in a culture to test fitness against a green, fluorescent wild-type strain. The percentage of analyzed cells in the culture measured was those displaying green fluorescence. In theory, this should mean if a knockout has reduced the fitness of a strain of yeast, the fluorescent wild-type strain would have a higher percentage than the knockout strain. If the knockout does not decrease fitness, they would be roughly equal. This may not be so in results. Sources of error may include contamination and human error.
The results here are not expected in theory. The YGL138C knockout strain had a significant disadvantage and the fluorescent wild-type strain dominated the given result. Test 1 must have been contaminated with a faster growing organism than yeast that did not display a fluorescent behavior like the wild-type fluorescent yeast used. However, Strain 2 appears to follow an expected result and suggests the knockout strain had a significant disadvantage imposed to its fitness, given the wild-type strain outgrew it.
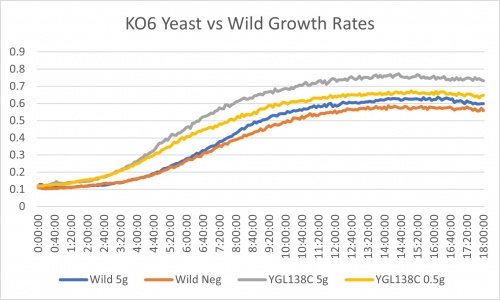
Ammonium Sulfate
YGL138C gene shows an interesting result. While the wild strain from 5g/ml of ammonium sulfate to none shows a 17.8% slower rate, it shows a 35.5% slower rate. For further analysis 0.5g/ml to no ammonium sulfate was compared too. Wild showed 1% slower rate and YGL138C showed 38% slowed rate. Based on the differences of these two treatments in the wild to the YGL138C KO strain it could be said that there was experimental error. The differences in percent are 16.8% in the wild and roughly 3% in the KO strain. The doubling time of the negative control is like the wild as well. It could be that errors in volume transfers occurred causing increased growth in the other treatments. It could also be that this strain with the knocked out YGL138C gene is in fact more sensitive to nitrogen starvation.
<protect>
UV Light
Results:
- Experiment 1:0sec=437 colonies, 600sec=81 colonies.
- Experiment 2:0sec=135 colonies, 600sec=28 colonies.
Interpretation: The two 0sec plates are on top and the 600sec plates are on the bottom. The 0sec yeast plate from experiment 2 had a lot of contamination along with the yeast while the 600sec plate from experiment 2 had very little. This yeast strain was about 80% for both plates which is more significant than some of the other strains however I'm not sure I'd say this is significant. It's possible that the number of yeast colonies was miscounted on the 0sec experiment 2 plate because of how small many of colonies are and how much contamination there is. Though it may be possible the gene played a role in UV resistance, it's hard to say with only one run of the experiment being good.
Calcofluor White
From this data we can determine that there was moderate growth inhibition on this strain of yeast. The modified yeast had a doubling rate that was about twenty minutes slower than the wild type on average, and when compared to the effect on other yeast strains this is a moderate effect on the inhibition of growth.
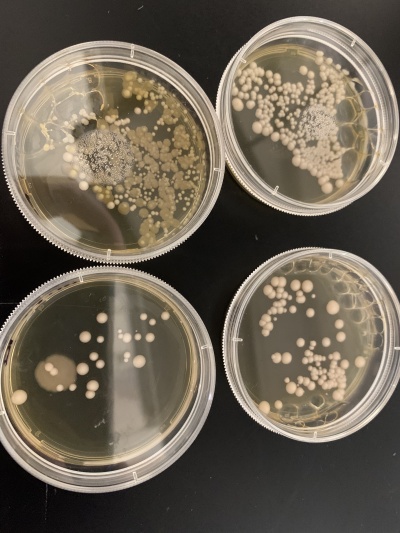
Heat Shock
The yeast strain YGL138C was impacted significantly by the heat shocking of the thermal cycler. As seen in the image, the controlled plate grew a lot of small DNA colonies all throughout the majority of pitri dish. It looks like a very thick sheet of cells. When compared to the two trial heat shocking plates, there are a lot less cells being grown. The trial cells are slightly larger than those of the controlled, but not by much. The quantity of the cells is much smaller, and the quality (size) of the cells is not that much bigger than what they're being compared to. This gene was heat sensitive, but not as much as the YLR426W yeast strain.
Caffeine Group 2
Doubling Time at 0.25 concentration
YGL138C Td = (217-1) x ln(2)/ln(0.2/0.1) = 216 minutes; YGL138C Td= (153-33) x ln(2)/ln(.7/.1)= 42 minutes; Wild Type Td = (146-66) x ln(2)/ln(.6/.2) = 88 minutes These doubling times show that the 0.25 mM concentration in YGL138C slowed down the rate of growth. The caffeine was able to slow down metabolic activity. Caffeine may effect the function of the Rad52 protein that aids in DNA double-stranded breaks and maintaining genome stability. The caffeine may also aid in the process of the cell protecting itself from reacting oxygen species. Since the concentration is small in volume, apoptosis was not induced for cell function.
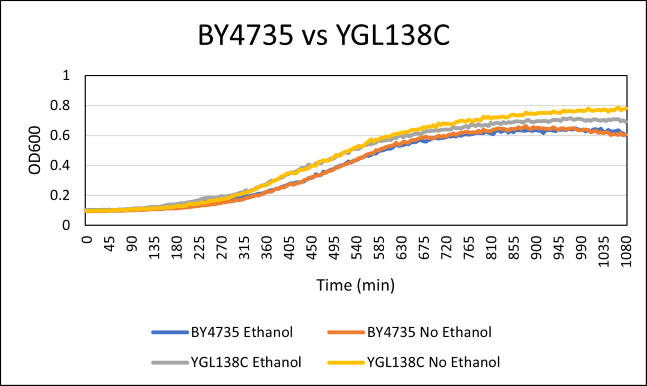
Ethanol Sensitivity
The YGL138C yeast strain was determined to be barely different in growth to the BY4735 strain. The YGL138C strain had a doubling time of 191 minutes while the BY4735 strain has a doubling time of 182 minutes. This means that the modification had little to no effect on the way that the cell interacts with ethanol. The doubling time of the BY4735 with ethanol had a doubling time of 218 minutes, while the YGL138C with ethanol had a doubling time of 212 minutes.
[[1]]
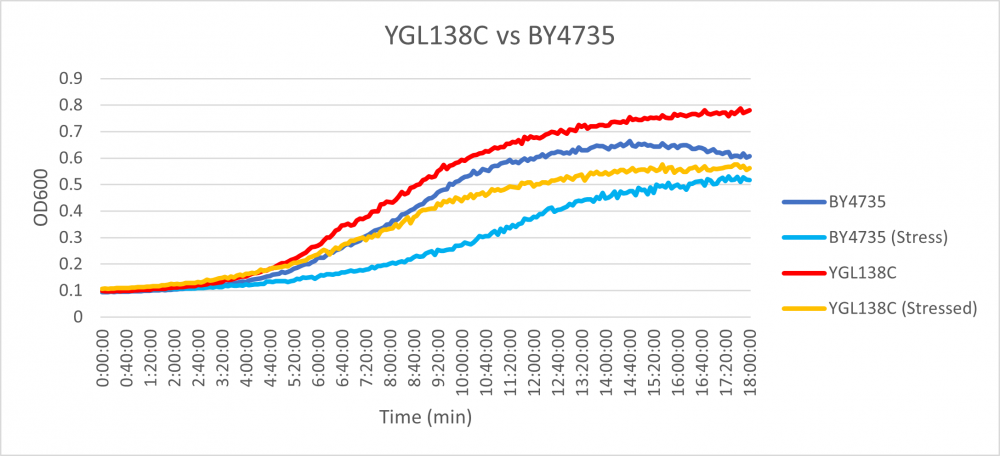
Hyrdoxyurea
The YGL138X strain had an average doubling time that was very similar to the growth rate of the BY4735 strain. The stressed YGL138C strain had a doubling time of 286 minutes, while the average doubling time of the stressed BY4735 strain had an average doubling time of 278 minutes. What is similar in both unstressed samples is what is interesting, the unstressed YGL138C strain had an average doubling time of 167 minutes, while the BY4735 unstressed strain had an average doubling time of 182 minutes. This means the stressor had little effect on the rate of growth of the strain. [2]
References
See Help:References on how to add references
See Help:Categories on how to add the wiki page for this gene to a Category </protect>