YBL113C
Share your knowledge...Edit this entry! <protect>
| Systematic name | YBL113C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr II:2658..280 |
| Primary SGDID | S000002153 |
Description of YBL113C: Helicase-like protein encoded within the telomeric Y' element[1]
</protect>
Contents
- 1 Community Commentary
- 2 References
- 3 UW-Stout/Nonionic Detergent SP22
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
UW Stout/D2O SP22
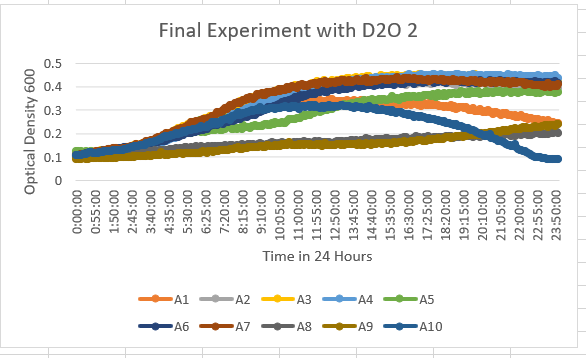
In the results of the 35% dilution of D2O with the Knock-out Yeast Strain YBL113C (A9) we can see that the growth curve stayed in the 0.1-0.3 range throughout the data collected in 24 hours. This tells us it was more sensitive to the dilution of D2O.
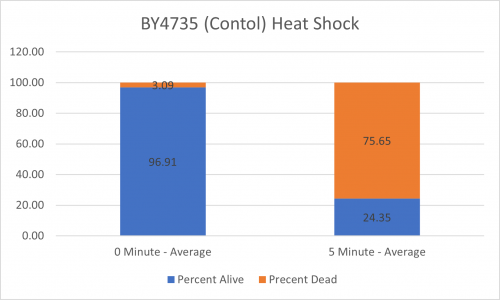
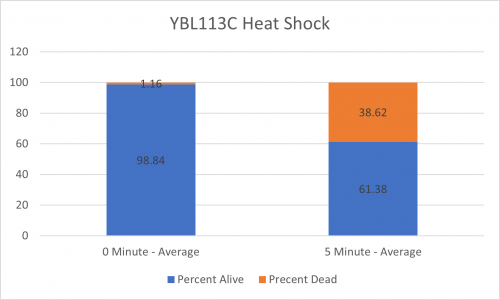
UW-Stout/Heat Shock SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to heat shock.
Results
Interpretation
From the data gathered, there was a medium positive effect to knocking out this gene when it came to the yeast's ability to hold up to heat shock. The modified yeast cells were 252% more resistant to heat shock than the control.
UW Stout/Sucrose Fermentation SP22
| Gene | Glucose | Fructose | Ethanol |
| Standard solution | 2.0000 | 0.2000 | 2.0000 |
| YDl109C | 0.3800 | 0.3933 | 0.3430 |
| YGL140C | 0.2212 | 0.2685 | 0.1867 |
| YOR111W | 0.3332 | 0.3598 | 0.1343 |
| YHL029C | 0.3870 | 0.2368 | 0.1151 |
| YDR307W | 0.4366 | 0.2487 | 0.0606 |
| YNL058C | 0.2710 | 0.3056 | 0.1577 |
| YCL049C | 0.4078 | 0.3052 | 0.1969 |
| YGR079W | 0.4042 | 0.1589 | 0.0080 |
| YBL113C | 0.3498 | 0.2012 | 0.1434 |
| BY4735 | 0.3171 | 0.3084 | 0.3541 |
Interpretation
This gene produced 40.50% of the amount of ethanol that the wild type produced.
UW-Stout/Caffeine SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to 4mM of caffeine following this protocol.
Results
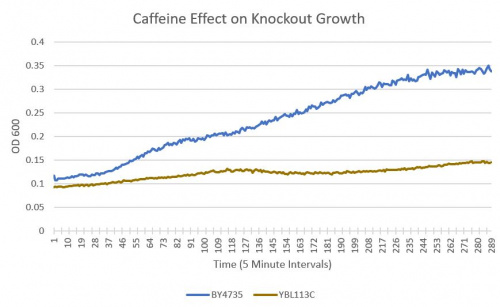
- BY4735(wild type yeast) and YBL113C(knockout yeast gene) growth after being subjected to 4mM caffeine solution.
Interpretation
As can be seen in the growth chart, YBL113 shows a much slower growth rate compared to the Wild-Type yeast.
Interpretation
From the growth curve, we can see a clear difference between the rates at which the Wild-Type and YBL113C showed growth. At the same concentration of 4mM caffeine, YBL113 showed greatly reduced growth implying a greater sensitivity to caffeine.
UW-Stout/Hydrogen Peroxide SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to hydrogen peroxide.
Results
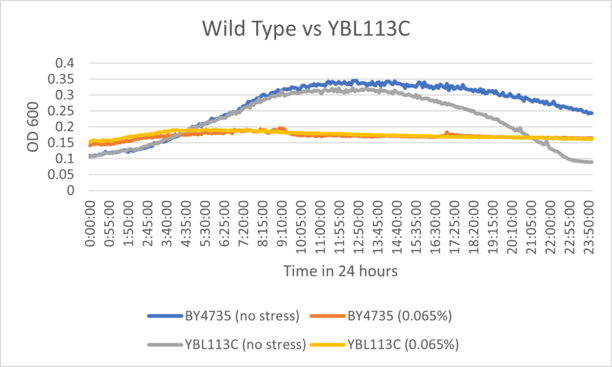
BY4735 and YBL113C after being exposed to 0.065% dilution of hydrogen peroxide solution.
Interpretation
From the growth curve, we can see that the knockout strain followed a similar sensitivity pattern to the wild type strain, suggesting that the grow has inhibited identically.
UW Stout/Nystatin SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to 1µg/ml Nystatin solution.
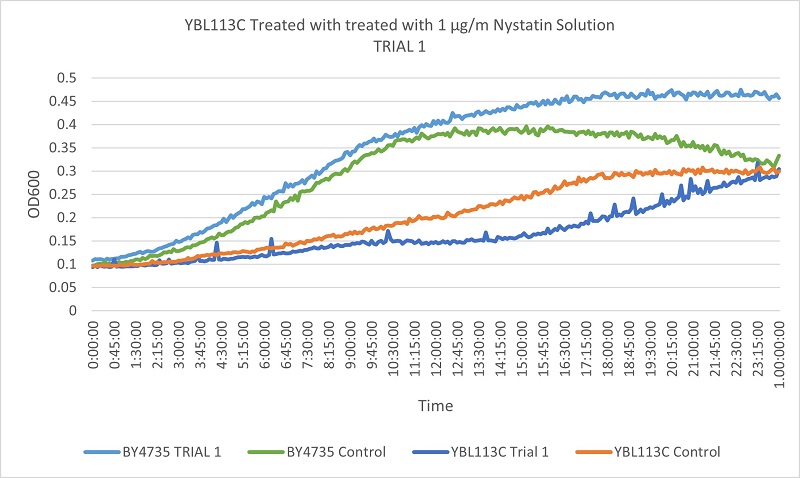
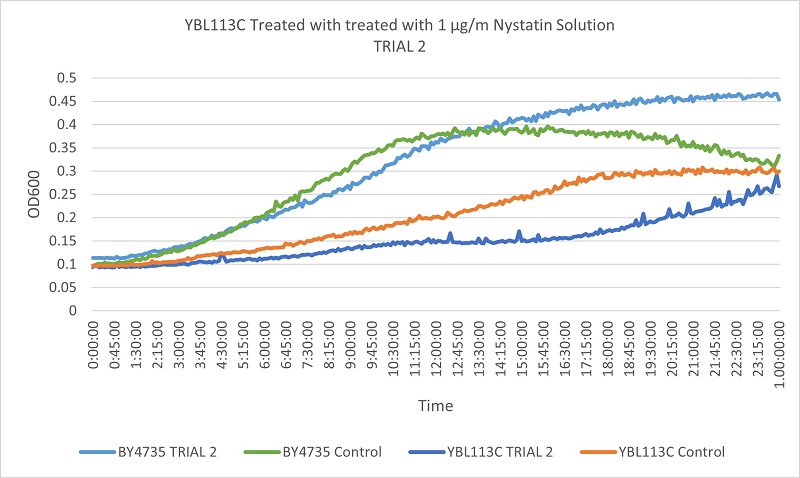
Results
Based on the growth curves, we can see that the YBL113C strain is much more slowly growing, so the curves are much lower. However, the differences between the control and experimental line in the wild type yeast and the YBL113C strain are very similar, meaning cutting out YBL113C does not greatly effect its resistance to Nystatin.
UW-Stout/Formamide SP22
Results
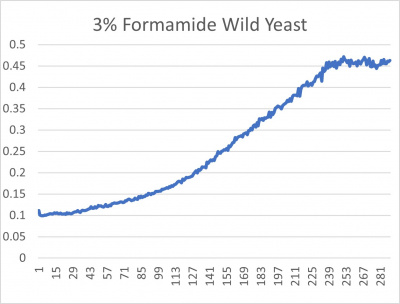
- 3% formamide line graph: x-axis= growing time (min); y-axis= optical density 600
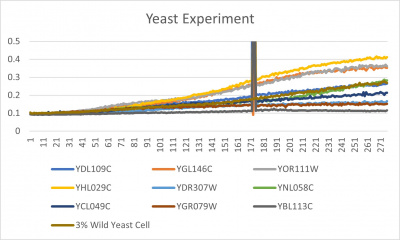
- Trial 2 line graph: x-axis= growing time (min); y-axis= optical density 600; key= yeast strains and corresponding color to the individual lines in the line graph
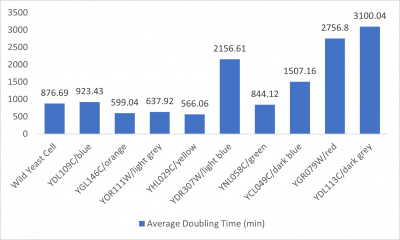
- Average doubling time bar graph: x-axis= yeast strains and corresponding colors to the trial 2 line graph; y-axis= average doubling time (min)
Interpretation
The wild yeast cell treated with 3% formamide had a doubling time of 876.69 min. The 3% formamide solution added to nine transformed yeast cells was then compared to the wild yeast cell using the computed doubling times to see the effects of the formamide. The results from the graph and table were not completely expected. YBL113C had the slowest growth rates, with a doubling time of 3100.04 min. Where the 3% formamide did not seem to hinder the growth process of some of the strains, it had the adverse effect with the YBL113C strain slowing down the growing process substantially. Therefore, it can be concluded that the transformed yeast cells that grew substantially slower in comparison to the wild yeast cell had a unique response to the stress of the formamide used in the experiment.
References
See Help:References on how to add references
See Help:Categories on how to add the wiki page for this gene to a Category </protect>
UW-Stout/Nonionic Detergent SP22
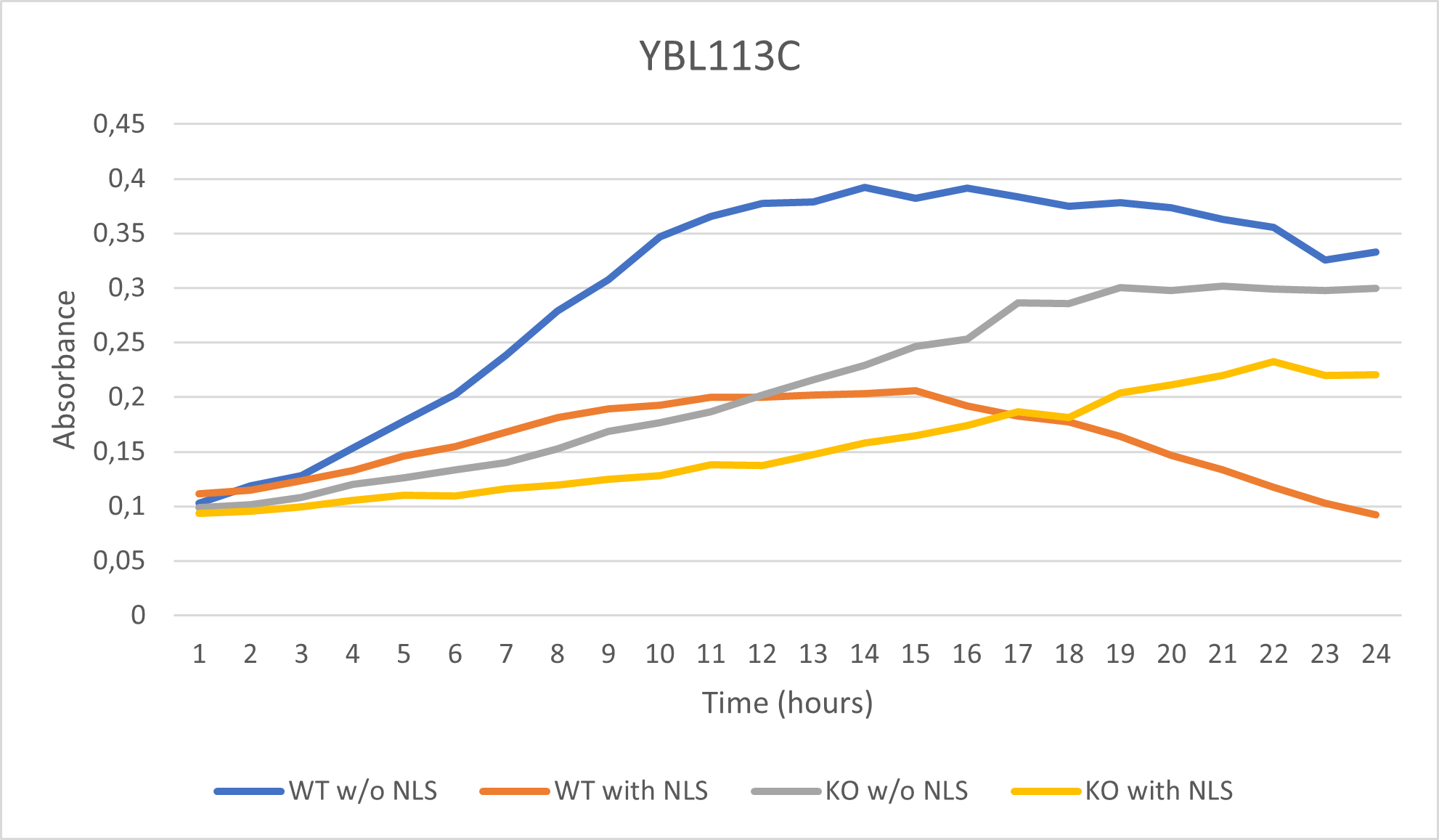
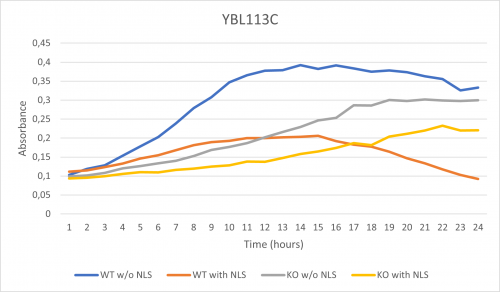
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to 0.001% concentrated NLS. (Nonionic detergent)
Interpretation:
-According to the graph above, we can see that the KO yeast cell YBL113C follow a smaller sensitivity to the stress than the Wild type strain.
-We can then conclude that the knocked-out gene in YBL113C doesn't have a significant impact on the growth of the yeast cell.
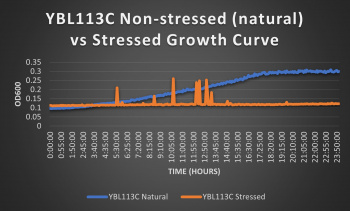 Knocked out gene seems to have a very significant effect on cell growth in pH 7 environment based on the flattening of the growth curve. Spikes in stressed line are to be ignored; they occurred due to clumping of cells during analysis. We are focusing on the general linear trend of the growth curve. Please see protocol for specific quantitative doubling time results.
Knocked out gene seems to have a very significant effect on cell growth in pH 7 environment based on the flattening of the growth curve. Spikes in stressed line are to be ignored; they occurred due to clumping of cells during analysis. We are focusing on the general linear trend of the growth curve. Please see protocol for specific quantitative doubling time results.
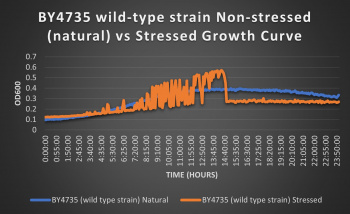 The wild-type strain was used as a control in this experiment; no genes were knocked out, it just represents how the experiment ran on the knocked-out gene strains affects a "typical" cell growth curve. Inclusion of this data on each knock-out strain growth curve graph made for difficult interpretation, so it has been separated. Please refer to this graph as a control when viewing the knock-out strain growth curve graph. Spikes in stressed line are to be ignored; they occurred due to clumping of cells during analysis. We are focusing on the general linear trend of the growth curve.
The wild-type strain was used as a control in this experiment; no genes were knocked out, it just represents how the experiment ran on the knocked-out gene strains affects a "typical" cell growth curve. Inclusion of this data on each knock-out strain growth curve graph made for difficult interpretation, so it has been separated. Please refer to this graph as a control when viewing the knock-out strain growth curve graph. Spikes in stressed line are to be ignored; they occurred due to clumping of cells during analysis. We are focusing on the general linear trend of the growth curve.