SGD Quarterly Newsletter, Summer 2014
About this newsletter:
This is the Summer 2014 issue of the quarterly SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community.
You can also subscribe to SGD's RSS feed to receive updates on SGD news:
http://www.yeastgenome.org/feed
Contents
SGD at the Yeast Genetics Meeting
SGD staff will be hosting a workshop, posters, and an exhibit table at the GSA Yeast Genetics Meeting in Seattle to showcase the items mentioned below and more! At the workshop, Computational Tools at SGD, we will be discussing our powerful search tool, YeastMine, what's new in the realm of Strains and Sequences, and new displays in SGD. Bring your questions and comments - we love feedback!
Find these SGD staff members, as well as those presenting posters, at the Workshop and the Exhibit table:
Follow @yeastgenome and #YEAST14 on Twitter for the latest yeast research being presented at YGM.
Workshop: "Computational Tools at SGD"
Thursday, July 31, 1:30 - 3:00 PM
Kane Hall, Room 220
Featured topics: YeastMine (our powerful search tool), Sequences and Strains update, New data displays at SGD
Posters
In addition to the Workshop, SGD curators will present 4 posters - please stop by and chat with us!
| Poster | Date and Time | Poster Title | Presenter |
| 318C | Friday, August 1 7:30 - 8:30 PM HUB Grand Ballroom |
Defining the transcriptome of Saccharomyces cerevisiae | |
| 387C | Friday, August 1 8:30 - 9:30 PM HUB Grand Ballroom |
Yeast - it simply has a lot to say about human disease | |
| 411C | Friday, August 1 8:30 - 9:30 PM HUB Grand Ballroom |
Transcriptional regulation and protein complexes in budding yeast | |
| 459C | Friday, August 1 8:30 - 9:30 PM HUB Grand Ballroom |
Staying current and modern: Overhauling an actively-used model organism database website |
Exhibit Table
SGD will also have an exhibit table at the YGM. Come by to take a spin on our site, receive a prize for taking a survey, learn about various features of the database, and provide us with feedback as to what we can do to improve SGD! Look for us wearing our SuperBud fleece jackets, and feel free to flag any of us down!
SGD is getting more social
Subscribe to SGD’s YouTube Channel
Have questions about the GO Term Enrichment widget in YeastMine? Wondering what Educational resources are available at SGD? Learn more about and keep up-to-date with SGD’s many tools by viewing great tutorials at the Saccharomyces Genome Database Channel on YouTube. Visit and subscribe today!
Connect to us through LinkedIn, Facebook, and Twitter
SGD is on LinkedIn, Facebook, and Twitter! Follow SGD's pages to get updates on the latest developments at SGD and in the yeast community.
We have redesigned the Protein page to include a new tabular display of protein domains. This table provides the identifier for each domain and illustrates the respective locations of the domains within the protein. In addition to this new table, the domains are displayed in an interactive network diagram that presents the proteins that share these domains with your protein of interest (See image below, left).
Another new feature on the Protein page is the display of phosphorylation sites within the protein’s sequence (as curated by BioGRID). This feature is available for both the reference strain S288C and other commonly used S. cerevisiae strains, using the pull-down to select the desired strain view (See image below, right).
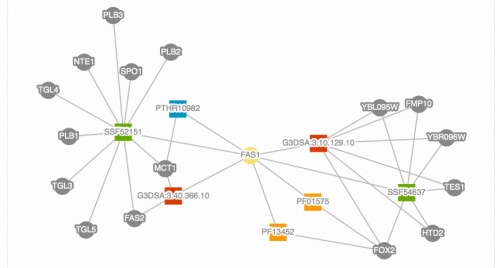 Proteins (gray circles) that share domains (colored squares) with Fas1p (yellow circle). |
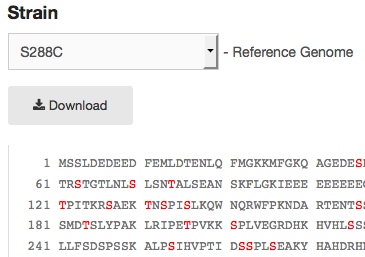 An example of some of the phosphorylation sites in Swe1p (red residues). |
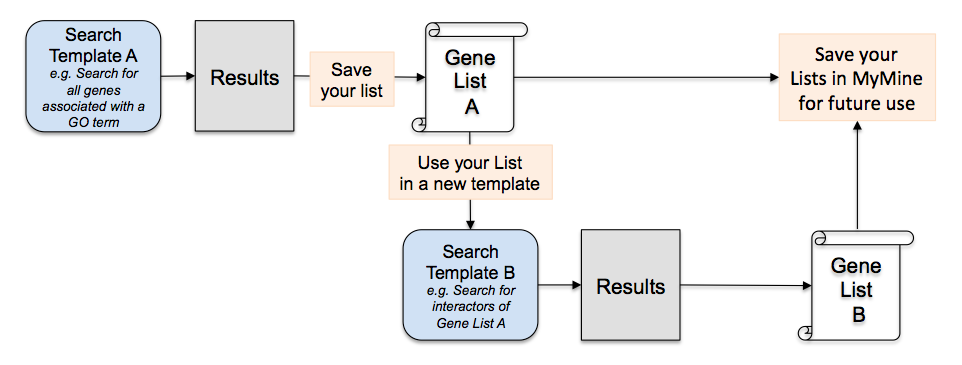
Create, Analyze, and Save Lists in YeastMine
If you love to make and analyze lists of genes, you will love YeastMine – you can use it to create all kinds of lists! For instance, use a YeastMine template to search for all genes associated with a given GO term or phenotype observable and save these genes as a list. Or, search for all genes that interact with your gene of interest and save that as a gene list.
What’s even more fun is that you can make lists from your lists! For instance, take the list of genes you found to be associated with a given GO term and plug it into a YeastMine query template to find all the genes that interact with your list of genes – then save those genes as a list. The possibilities are endless given the different types of queries you can perform using YeastMine! Who knows what biological connections you will uncover? Lastly, save your lists for future use by creating a MyMine account – all you need to sign up is an email and a password.
You can find a link to YeastMine in the top right corner of most SGD pages (“YeastMine: Batch Analysis or Advanced Search”) or go to SGD’s purple main menu bar, click on “Analyze” and select “Gene Lists” to go straight to creating a List in YeastMine.
To see how simple it is to save your search results as a List in YeastMine, view this brief tutorial – YeastMine: Saving Search Results as a List.
Educational Resources on the SGD Community Wiki
Did you know you can find and contribute teaching and other educational resources to SGD? We have updated our Educational Resources page, found on the SGD Community Wiki. There are links to teaching resources such as classroom materials, courses, and fun sites, as well as pointers to books, dedicated learning sites, and tutorials that can help you learn more about basic genetics.
We would like to encourage others to contribute additional teaching or general educational resources to this page. To do so, just request a wiki account by contacting us at the SGD Help Desk - you will then be able to edit the SGD Community Wiki. If you prefer, we would also be happy to assist you directly with these edits.
Note that there are many other types of information you can add to the SGD Community Wiki, including information about your favorite genes, protocols, upcoming meetings, and job postings. The Community Wiki can be accessed from most SGD pages by clicking on "Community" on the main menu bar and selecting "Wiki." The Educational Resources page is linked from the left menu bar under "Resources" from all the SGD Community Wiki pages. For more information, please view the video Educational Resources on the SGD Community Wiki.
Recent SGD Publications
1. Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, Dwight SS, Hitz BC, Karra K, Nash RS, Weng S, Wong ED, Lloyd P, Skrzypek MS, Miyasato SR, Simison M, Cherry JM (2013) The reference genome sequence of Saccharomyces cerevisiae: Then and now. Database (Oxford) G3 (Bethesda). 2014 Mar 20;4(3):389-98. doi: 10.1534/g3.113.008995. PMID:24374639
2. Engel SR, Cherry JM (2013) The new modern era of yeast genomics: community sequencing and the resulting annotation of multiple Saccharomyces cerevisiae strains at the Saccharomyces Genome Database. Database (Oxford) 2013 Mar 13;2013:bat012. doi: 10.1093/database/bat012. PMID:23487186
Upcoming Meetings
- Cold Spring Harbor Labs, Cold Spring Harbor, New York
- July 22 - August 11, 2014
- University of Washington, Seattle, Washington
- July 29 - August 3, 2014
- EMBL Conference
- Frontiers in Fungal Systems Biology
- EMBL Heidelberg, Germany
- September 28-30, 2014
- Vipava and Nova Gorica, Slovenia
- October 9-12, 2014
- EMBL Conference
- Experimental Approaches to Evolution and Ecology using Yeast & other Model Systems
- EMBL Heidelberg, Germany
- October 12-15, 2014
*asterisks indicate attendance by SGD
Note: If you no longer wish to receive this newsletter, please contact the SGD Help Desk at sgd-helpdesk@lists.stanford.edu.