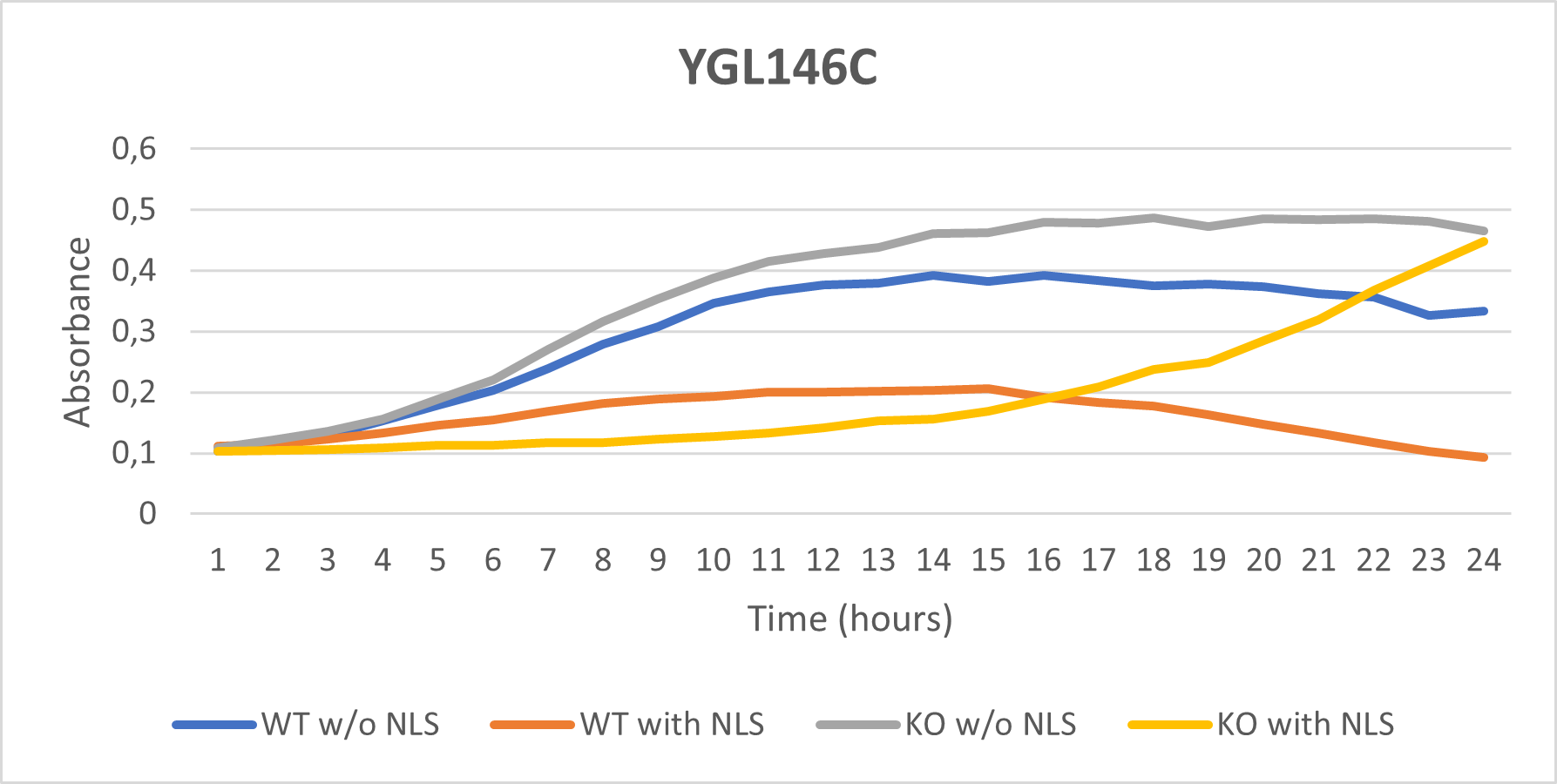
YGL146C
Share your knowledge...Edit this entry! <protect>
| Systematic name | YGL146C |
| Gene name | RRT6 |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr VII:229686..228751 |
| Primary SGDID | S000003114 |
Description of YGL146C: Putative protein of unknown function; non-essential gene identified in a screen for mutants with increased levels of rDNA transcription; contains two putative transmembrane spans, but no significant homology to other known proteins[1][2][3]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
UW Stout/D2O SP22
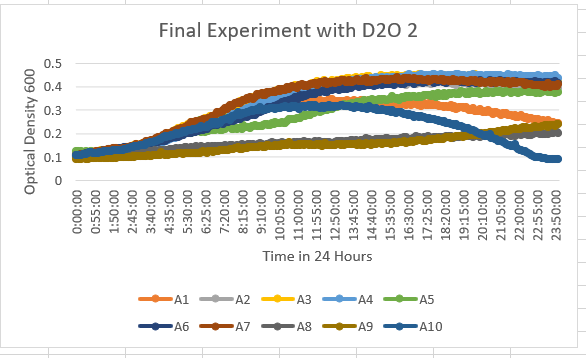
The YGL146C Knock Out Yeast Strain was A2 and definitely unaffected by the 35% D20 dilution. It showed when it stayed in a maximum range of 0.4-0.5 OD 600.
UW Stout/Sucrose Fermentation SP22
| Gene | Glucose | Fructose | Ethanol |
| Standard solution | 2.0000 | 0.2000 | 2.0000 |
| YDl109C | 0.3800 | 0.3933 | 0.3430 |
| YGL140C | 0.2212 | 0.2685 | 0.1867 |
| YOR111W | 0.3332 | 0.3598 | 0.1343 |
| YHL029C | 0.3870 | 0.2368 | 0.1151 |
| YDR307W | 0.4366 | 0.2487 | 0.0606 |
| YNL058C | 0.2710 | 0.3056 | 0.1577 |
| YCL049C | |||
| YGR079W | |||
| YBL113C | 0.3498 | 0.2012 | 0.1434 |
| BY4735 | 0.3171 | 0.3084 | 0.3541 |
Interpretation
This gene produced 52.73% of the amount of ethanol that the wild type produced.
<protect>
UW-Stout/UV Light SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested under a UV Light using this protocol.
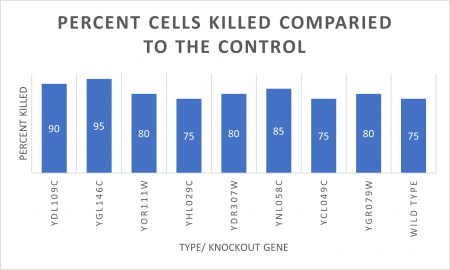
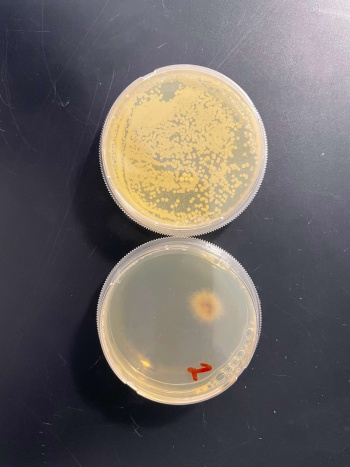
RESULTS
INTERPERTATION
In the graph and photos above, exposing this gene to 600 seconds of 400 Watt UV Light killed approximately 95% of yeast cell cultures, compared to its control counterpart, which was the same gene and amount of cells, just was not exposed to UV Light.
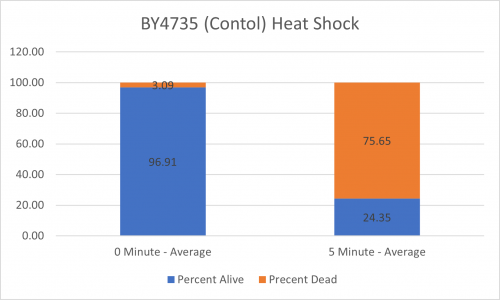
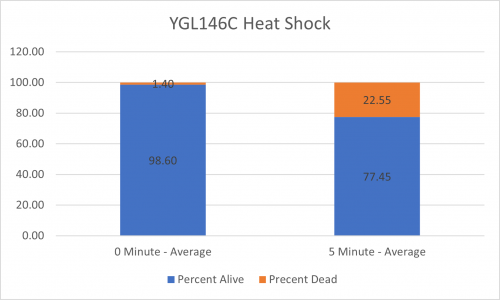
UW-Stout/Heat Shock SP22
As part of the University of Wisconsin Stout Orphan Gene Project this gene was tested by exposing the cells to heat shock.
Results
Interpretation
From the data gathered, there was a large positive effect to knocking out this gene when it came to the yeast's ability to hold up to heat shock. The modified yeast cells were 318% more resistant to heat shock than the control.
References
See Help:References on how to add references
- ↑ Hontz RD, et al. (2009) Genetic Identification of Factors That Modulate Ribosomal DNA Transcription in Saccharomyces cerevisiae. Genetics 182(1):105-19 SGD PMID 19270272
- ↑ Pearson BM and Schweizer M (2002) Basic functional analysis of six unknown open reading frames from Saccharomyces cerevisiae: four from chromosome VII and two from chromosome XV. Yeast 19(2):123-9 SGD PMID 11788967
- ↑ Voet M, et al. (1997) The sequence of a nearly unclonable 22.8 kb segment on the left arm chromosome VII from Saccharomyces cerevisiae reveals ARO2, RPL9A, TIP1, MRF1 genes and six new open reading frames. Yeast 13(2):177-82 SGD PMID 9046099
See Help:Categories on how to add the wiki page for this gene to a Category </protect>