Difference between revisions of "YAL061W"
(→Community Commentary) |
(→Zone of Inhibition of 30% Hydrogen Peroxide on YAL061W) |
||
| (21 intermediate revisions by the same user not shown) | |||
| Line 78: | Line 78: | ||
| − | === | + | ===[[UW Stout/Proline FA19|Growth in Proline and Ammonium Sulfate]]=== |
[[File:KO1.png|650 px]] | [[File:KO1.png|650 px]] | ||
| + | The YAL061W strand grew faster in ammonium sulfate media with an average doubling time of 35.43 minutes. When grown in proline, the average doubling time was 39.83 minutes. The wild types were close in time, but it grew faster in proline media with an average doubling time of 46.23. The average doubling time of the wild type in ammonium sulfate was 48.33 minutes. | ||
| − | ===Formamide Yeast Torture[[https://wiki.yeastgenome.org/index.php/UW_Stout/Formamide_FA19]]=== | + | ===1% Formamide Yeast Torture[[https://wiki.yeastgenome.org/index.php/UW_Stout/Formamide_FA19]]=== |
| − | [[ | + | [[Image:ko_strand_-1_finished.png]] |
| + | This graph shows the growth curve of the knockout yeast strain YAL061W and wild type BY4735 in 1% formamide in 3 trials. The average calculated doubling time for YAL061W with 1% formamide was 5,177.34 minutes, while the unstressed strain was 2,371.77 minutes. This concluded that the stressed out strain is highly sensitive to 1% formamide, and grows as a slower pace in this environment. Our stressed wild types doubling time was 3,178.81 minutes, and the unstressed was 5,887.54 minutes. The stressed wild type in this experiment proved to show better growth concluding that the addition of 1% formamide for this strain does not cause sensitivity. | ||
| + | |||
| + | [[File:Nitrogen_Starvation_YAL061W.jpg|463 x 599|Caption]] | ||
| + | |||
| + | The Wild Type BY4735 has data located at the Nitrogen Starvation Protocol. | ||
| + | ==[[UW-Stout/Nitrogen_starvation_FA19|Nitrogen starvation]]== | ||
<!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | <!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | ||
<!-- | <!-- | ||
| Line 94: | Line 101: | ||
--> | --> | ||
| − | + | == [https://wiki.yeastgenome.org/index.php?title=UW-Stout/Hydroxyurea_FA19&oldid=401696 Hydroxyurea] == | |
| + | [[File:AvaYAL061W.png]] | ||
| + | The averaging doubling time for YAL061W is 716.41 minutes. The averaging doubling time for the unstressed YAL061W is 583.31 minutes | ||
<!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | <!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | ||
| Line 103: | Line 112: | ||
--> | --> | ||
| + | ===Zone of Inhibition of 30% [https://wiki.yeastgenome.org/index.php/UW-Stout/Hydrogen_peroxide_FA19 Hydrogen Peroxide] on YAL061W=== | ||
| + | [[File:JaredJ2.PNG]] | ||
| + | [[File:JJ2.PNG]] | ||
| − | + | The bar graph represents the zone of inhibition of the knockout strain versus the wild type. Our data collected shows that the knockout strain saw a larger zone of inhibition (cells were not able to grow there) than the wild type. The error bar on the knockout strain represents the standard deviation of the experiment and how recreatable our results were with these results. From these measurements I can say that YAL061W undergoes more stress from the oxidative properties of 30 percent hydrogen peroxide. The zone of inhibition is larger because the yeast cells are not able to grow as well as the wild type can in these conditions. For the strain YAL061W our plate values were as follows (in cm) 2.3, 2.4 and 2.33 compared to our wild type plate which stayed constant 2.1, 2.1 and 2.2 cm | |
| − | |||
| − | |||
<protect> | <protect> | ||
Latest revision as of 08:36, 18 December 2019
Share your knowledge...Edit this entry!
<protect>
| Systematic name | YAL061W |
| Gene name | BDH2 |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr I:33448..34701 |
| Primary SGDID | S000000057 |
Description of YAL061W: Putative medium-chain alcohol dehydrogenase with similarity to BDH1; transcription induced by constitutively active PDR1 and PDR3[1][2][3][4]
</protect>
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
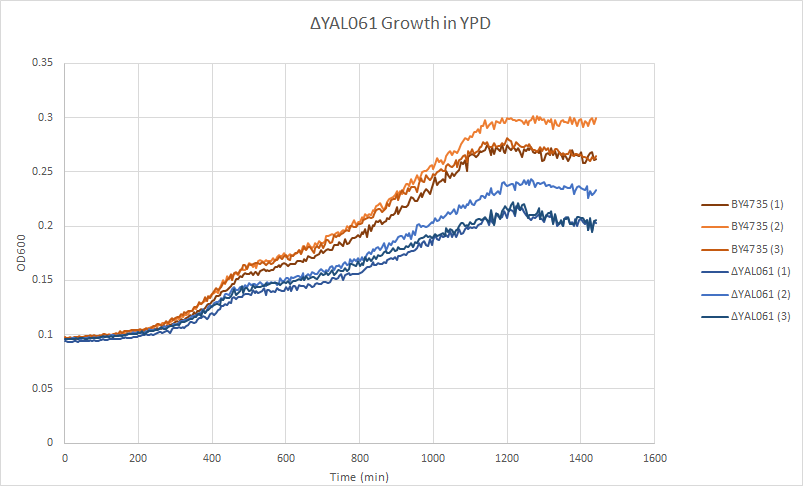
Growth Curve
In a BY4735 background, knocking out YAL061W seems to have a moderate effect on growth rate in log-phase. In this assay, the BY4735 strain's doubling time was 361 minutes, while the YAL061 knock-out strain's doubling time was 469 minutes. (These doubling times are the means of three experiments.)
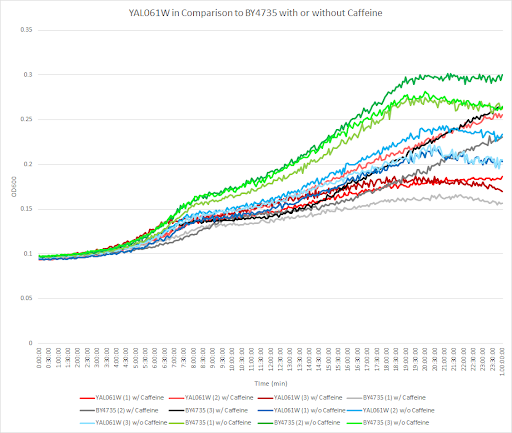
Caffeine and Yeast Cells
This graph shows the growth curve with and without caffeine. It has the knockout strain and wild type yeast cells. The caffeine enhanced the growth in both wild type and knockout strain. The average doubling time between the three experiments for the knockout strand was 777.27 minutes. The caffeine also had a positive effect on the growth curve for the wild type with the average doubling time being 1245.02 minutes. The results were concluded from the wild type and knockout strand was with this amount of caffeine it leads to an increase in growth.
The protocol can be found at
Caffeine
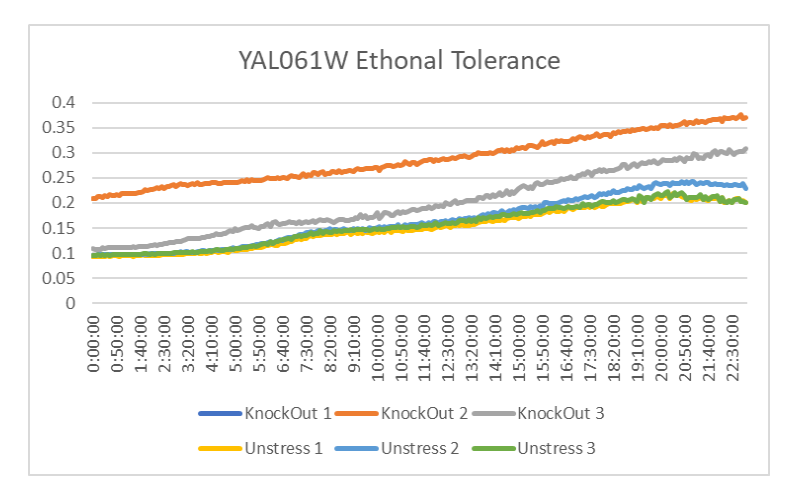
Ethanol Tolerance for YAL061
This graph shows the growth curve of ethanol tolerance in YAL061. The unstressed lines represent YAL061 that was not treated with ethanol. If you examine the graph you will notice that the strands that were treated with ethanol grew more than the ones that weren't. The average doubling time for the stressed yeast was 1263.28 minutes. The average doubling time for the unstressed yeast was 1174.42 minutes. The results show that YAL061 in ethanol leads to an increase in growth.
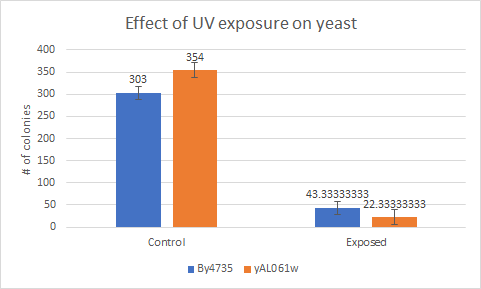
This bar graph shows us that compared to a BY4735 background, Knocking out yAL061w makes yeast more sensitive to UV light. The protocol can be found at
UV LIght Exposure
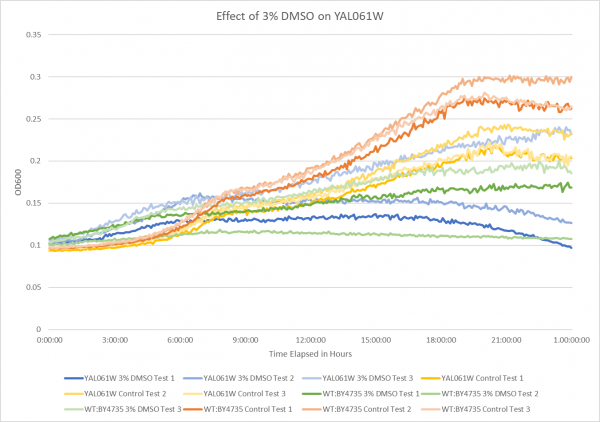
Effect of 3% DMSO on YAL061W
This graph represents the growth curve of DMSO tolerance in YAL061W. When calculated out the average doubling time for YAL061W with 3% DMSO equaled 906 minutes. The average doubling time for YAL061W without 3% DMSO equaled 683 minutes. This showed that 3% DMSO was in fact stressing out the knockout strain YAL061W. If we compare these average doubling times with those of WT:BY4735 which are 1529 minutes with 3% DMSO, and 514 minutes without DMSO it can be determined that by knocking out the YAL061W gene the doubling time increased slightly without DMSO. With adding DMSO the yeast grew faster without having the YAL061W gene.
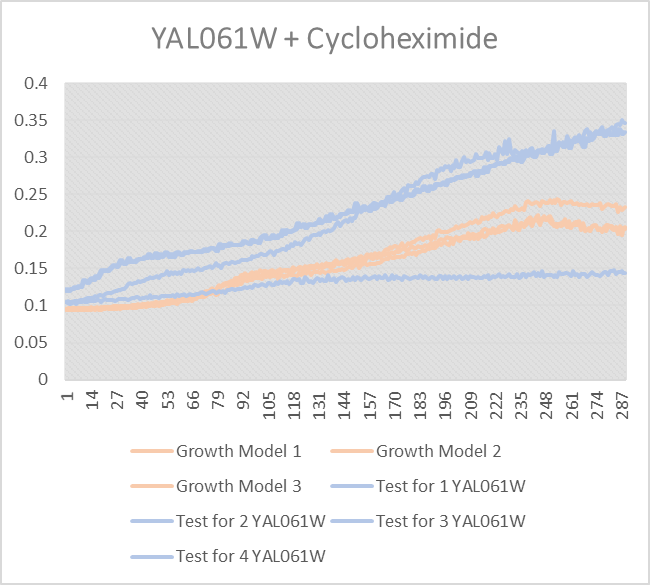
Effect of Cycloheximide on YAL0061W
This was Cycloheximide with the first KO Gene. The Growth Models 1-3 are the KO not being distressed. Tests 1-4 were our results. Most of the yeast survived or even did better,except for test 1, which is weird. And we are unsure why.
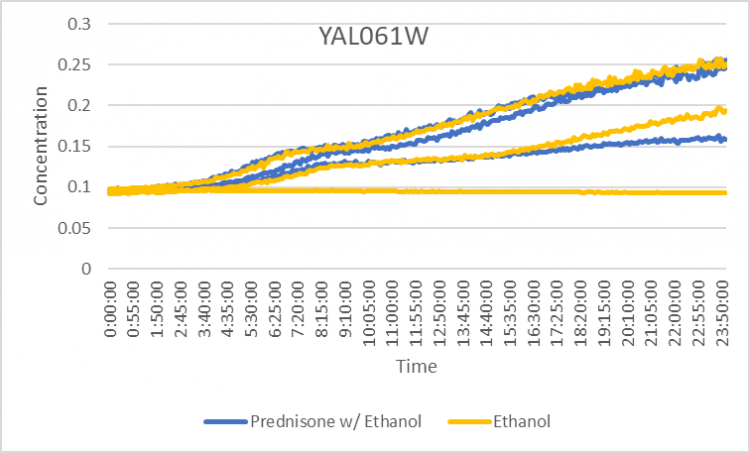
Prednisone Effects on YAL061W
The graph above shows how dilutions P2 (30 µg/ml) and E2 (10% ethanol solution) impacted the growth rate of the knockout yeast strain YAL061W over 24 hours. The concentration did not double for P2 or E2 during test 3 for this experiment. Taking only the first two test results for each, the doubling rate averages to around 1182.5 minutes for ethanol only. As for the Prednisone with ethanol, the doubling rate averages to around 962.5 minutes. Overall, ethanol decreased the rate of growth by almost 220 minutes. These results conclude that Prednisone promotes the growth of yeast cells without the YAL061W gene.
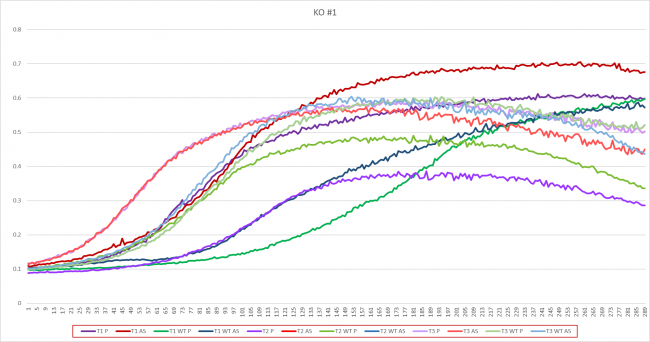
Growth in Proline and Ammonium Sulfate
The YAL061W strand grew faster in ammonium sulfate media with an average doubling time of 35.43 minutes. When grown in proline, the average doubling time was 39.83 minutes. The wild types were close in time, but it grew faster in proline media with an average doubling time of 46.23. The average doubling time of the wild type in ammonium sulfate was 48.33 minutes.
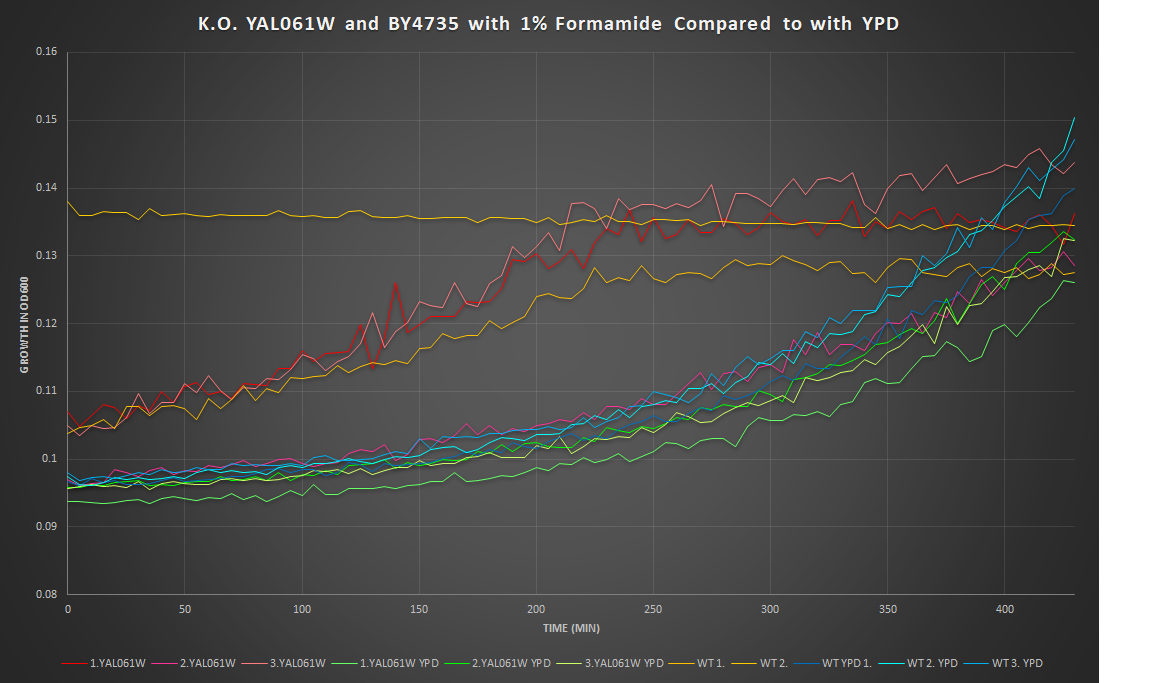
1% Formamide Yeast Torture[[1]]
This graph shows the growth curve of the knockout yeast strain YAL061W and wild type BY4735 in 1% formamide in 3 trials. The average calculated doubling time for YAL061W with 1% formamide was 5,177.34 minutes, while the unstressed strain was 2,371.77 minutes. This concluded that the stressed out strain is highly sensitive to 1% formamide, and grows as a slower pace in this environment. Our stressed wild types doubling time was 3,178.81 minutes, and the unstressed was 5,887.54 minutes. The stressed wild type in this experiment proved to show better growth concluding that the addition of 1% formamide for this strain does not cause sensitivity.
The Wild Type BY4735 has data located at the Nitrogen Starvation Protocol.
Nitrogen starvation
Hydroxyurea
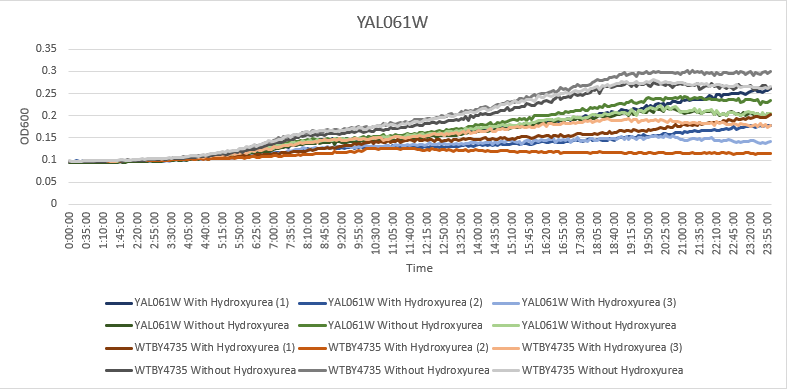 The averaging doubling time for YAL061W is 716.41 minutes. The averaging doubling time for the unstressed YAL061W is 583.31 minutes
The averaging doubling time for YAL061W is 716.41 minutes. The averaging doubling time for the unstressed YAL061W is 583.31 minutes
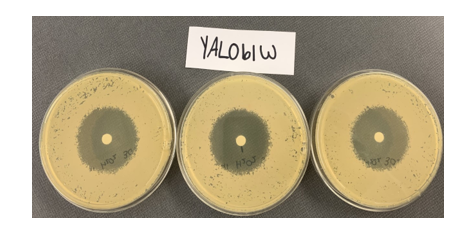
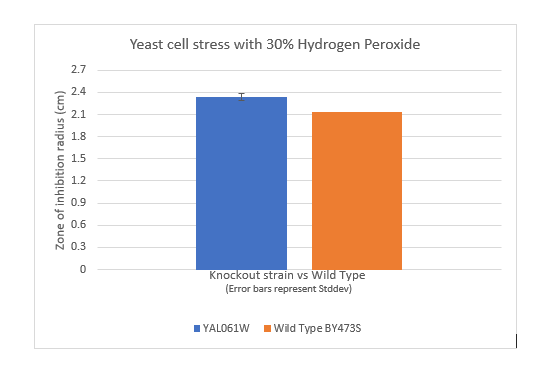
Zone of Inhibition of 30% Hydrogen Peroxide on YAL061W
The bar graph represents the zone of inhibition of the knockout strain versus the wild type. Our data collected shows that the knockout strain saw a larger zone of inhibition (cells were not able to grow there) than the wild type. The error bar on the knockout strain represents the standard deviation of the experiment and how recreatable our results were with these results. From these measurements I can say that YAL061W undergoes more stress from the oxidative properties of 30 percent hydrogen peroxide. The zone of inhibition is larger because the yeast cells are not able to grow as well as the wild type can in these conditions. For the strain YAL061W our plate values were as follows (in cm) 2.3, 2.4 and 2.33 compared to our wild type plate which stayed constant 2.1, 2.1 and 2.2 cm
<protect>
References
See Help:References on how to add references
- ↑ Devaux F, et al. (2002) Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett 515(1-3):25-8 SGD PMID 11943188
- ↑ Dickinson JR, et al. (2003) The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J Biol Chem 278(10):8028-34 SGD PMID 12499363
- ↑ Gonzalez E, et al. (2000) Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene product. Disruption and induction of the gene. J Biol Chem 275(46):35876-85 SGD PMID 10938079
- ↑ do Valle Matta MA, et al. (2001) Novel target genes of the yeast regulator Pdr1p: a contribution of the TPO1 gene in resistance to quinidine and other drugs. Gene 272(1-2):111-9 SGD PMID 11470516
See Help:Categories on how to add the wiki page for this gene to a Category </protect>