YKL121W
Share your knowledge...Edit this entry! <protect>
| Systematic name | YKL121W |
| Gene name | DGR2 |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr XI:214142..216700 |
| Primary SGDID | S000001604 |
Description of YKL121W: Protein of unknown function; null mutant is resistant to 2-deoxy-D-glucose and displays abnormally elongated buds[1][2][3]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
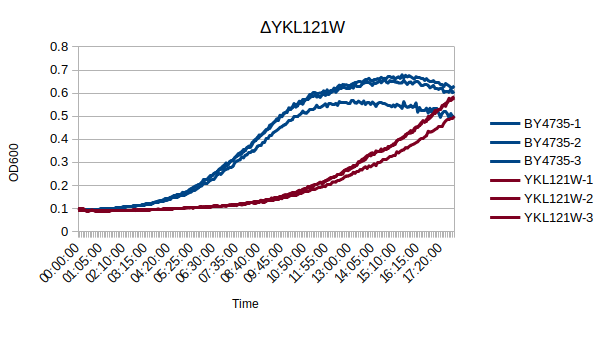
Growth Curve
In a BY4735 background, knocking out YKL121W seems to substantially slow the strain's growth. In this assay, the BY4735 strain's doubling time was 162 minutes, while the YKL121W knock-out strain's doubling time was 235 minutes. (These doubling times are the means of three experiments.)
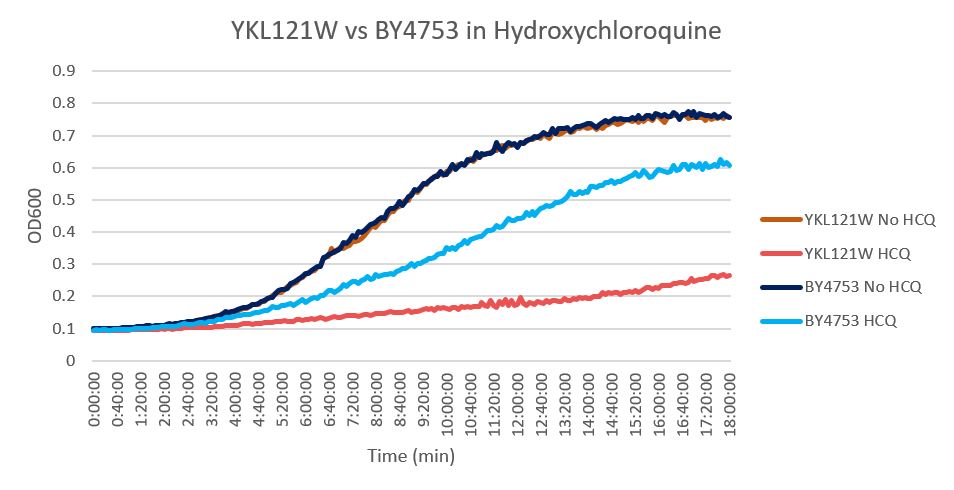
Hydroxychloroquine
HCQ refers to hydroxychloroquine
Under normal conditions, the BY4753 doubling time is 144 minutes, while that of YKL121W is 139 minutes. This proves that both of these strains grow at a similar rate. When grown in an environment with hydroxychloroquine, YKL121W doubled in 763 minutes, while BY4753 doubled in 276 minutes. Both strains are inhibited by hydroxychloroquine, but YKL121W growth is staggered much more. This is a possible indicator that the knocked-out gene was involved in cellular protection from hydroxychloroquine. (These doubling times and curves are the means of three experiments.)
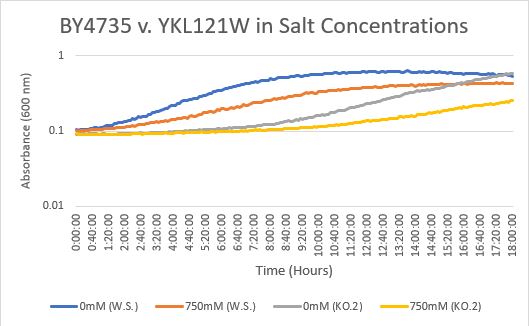
Salt Concentration (NaCl)
0mM NaCl conc. BY4735 strain's doubling time: 169 minutes
0mM NaCl conc. YKL121W strain's doubling time: 230 minutes
750mM NaCl conc. BY4735 strain's doubling time: 294 minutes
750mM NaCl conc. YKL121W strain's doubling time: 450 minutes
The graph above shows the growth rate for the previously listed strains and the relative level of NaCl concentration. Knocking out the gene negatively impacted the growth rate. Comparing the YKL121W doubling times in 0mM and 750mM NaCl, there is a tremendous effect on the growth rate. This effect negatively impacts the knockout strain's (YKL121W) growth rate.
Caffeine Group 1
Caffeine had an effect on the YKL121W strain. The doubling times are as followed:
Doubling time of YKL121W with caffeine: 406 minutes
Doubling time of YKL121W without caffeine: 358 minutes
Doubling time of BY4735 with caffeine: 138 minutes
Doubling time of BY4735 without caffeine: 187 minutes
According to the graph, the knockout strain's growth rate had significantly slowed down when caffeine was added. <protect>
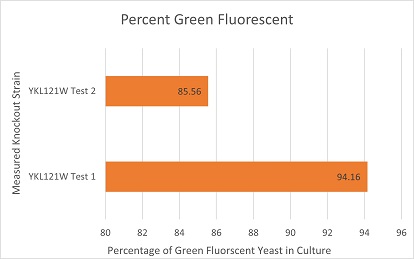
Competative Co-culture
These are the results of a competitive co-culture protocol. The knockout strain was grown in a culture to test fitness against a green, fluorescent wild-type strain. The percentage of analyzed cells in the culture measured was those displaying green fluorescence. In theory, this should mean if a knockout has reduced the fitness of a strain of yeast, the fluorescent wild-type strain would have a higher percentage than the knockout strain. If the knockout does not decrease fitness, they would be roughly equal. This may not be so in results. Sources of error may include contamination and human error.
The results here are expected in theory. The YKL121W knockout strain had a significant disadvantage and the fluorescent wild-type strain dominated as a result.
Ammonium Sulfate
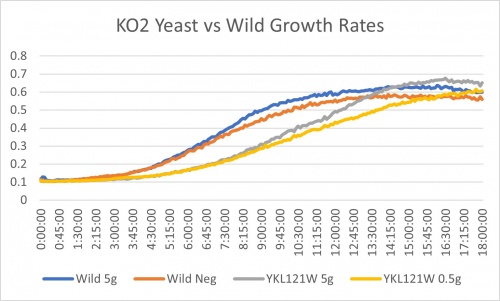
YKL121W gene does not indicate any difference from the wild strain when starving of nitrogen. We know this because in the wild it shows 17.8% slower growth when completely removing ammonium sulfate from the media and comparing that to the full amount of 5g/ml. While in YKL121W it shows 15.8%. These are more or less the same indicating that this gene does not make the yeast more or less sensitive to nitrogen starvation.
UV Light
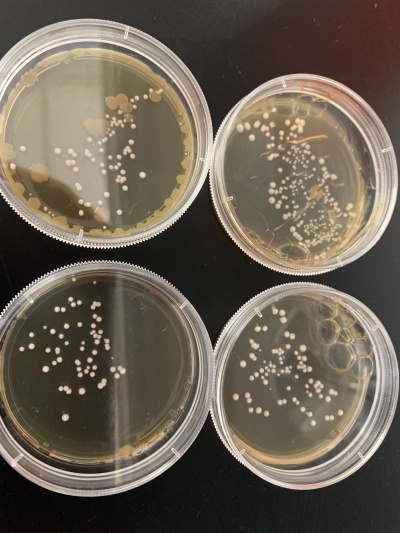
Results:
- Experiment 1:0sec=233 colonies, 600sec=86 colonies.
- Experiment 2:0sec=81 colonies, 600sec=60 colonies.
Interpretation: The two 0sec plates are on top and the 600sec plates are on the bottom. The two second experiment plates also had some contamination growing in the plate along with the yeast. The yeast didn't seem too affected by the UV light so I wouldn't say the Knock-out gene is related to UV light thought the contamination in experiment 2 makes me think something didn't quite go right.
=
References
See Help:References on how to add references
- ↑ Colleaux L, et al. (1992) Sequence of a segment of yeast chromosome XI identifies a new mitochondrial carrier, a new member of the G protein family, and a protein with the PAAKK motif of the H1 histones. Yeast 8(4):325-36 SGD PMID 1514329
- ↑ Ralser M, et al. (2008) A catabolic block does not sufficiently explain how 2-deoxy-D-glucose inhibits cell growth. Proc Natl Acad Sci U S A 105(46):17807-17811 SGD PMID 19004802
- ↑ Watanabe M, et al. (2009) Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Curr Genet 55(4):365-80 SGD PMID 19466415
See Help:Categories on how to add the wiki page for this gene to a Category </protect>