Difference between revisions of "YHL029C"
(→Community Commentary) |
|||
| Line 31: | Line 31: | ||
{{UW-Stout}} | {{UW-Stout}} | ||
| + | ===[[UW Stout/D2O SP 22]]=== | ||
| + | [[Image:4.28.2022 1.png]] | ||
| + | |||
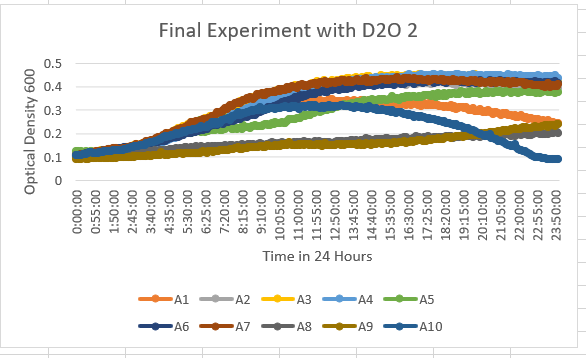
| + | The YHL029C Knock Out Yeast Strain (A4) is more 35% dilution D2O sensitive. With the maximum optical density of about 0.5-0.6 OD 600. | ||
| Line 43: | Line 47: | ||
<protect> | <protect> | ||
| + | |||
==References== | ==References== | ||
<!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | <!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | ||
{{RefHelp}} | {{RefHelp}} | ||
</protect> | </protect> | ||
Revision as of 09:24, 3 May 2022
Share your knowledge...Edit this entry! <protect>
| Systematic name | YHL029C |
| Gene name | OCA5 |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr VIII:47968..45929 |
| Primary SGDID | S000001021 |
Description of YHL029C: Cytoplasmic protein required for replication of Brome mosaic virus in S. cerevisiae, which is a model system for studying replication of positive-strand RNA viruses in their natural hosts[1][2]
</protect>
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
UW Stout/D2O SP 22
The YHL029C Knock Out Yeast Strain (A4) is more 35% dilution D2O sensitive. With the maximum optical density of about 0.5-0.6 OD 600.
<protect>
References
See Help:References on how to add references
- ↑ Huh WK, et al. (2003) Global analysis of protein localization in budding yeast. Nature 425(6959):686-91 SGD PMID 14562095
- ↑ Kushner DB, et al. (2003) Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A 100(26):15764-9 SGD PMID 14671320
See Help:Categories on how to add the wiki page for this gene to a Category </protect>