Difference between revisions of "YNL018C"
(→Caffeine Group 2) |
(→Hydroxychloroquine) |
||
| Line 40: | Line 40: | ||
===[[UW-Stout/Hydroxychloroquine_SP21|Hydroxychloroquine]]=== | ===[[UW-Stout/Hydroxychloroquine_SP21|Hydroxychloroquine]]=== | ||
| − | [[Image:YNL018C.png]] | + | [[Image:YNL018C-BY4735.png]] |
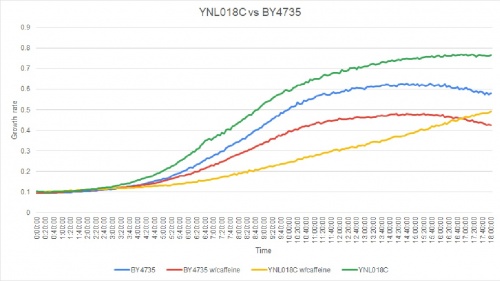
HCQ refers to hydroxychloroquine | HCQ refers to hydroxychloroquine | ||
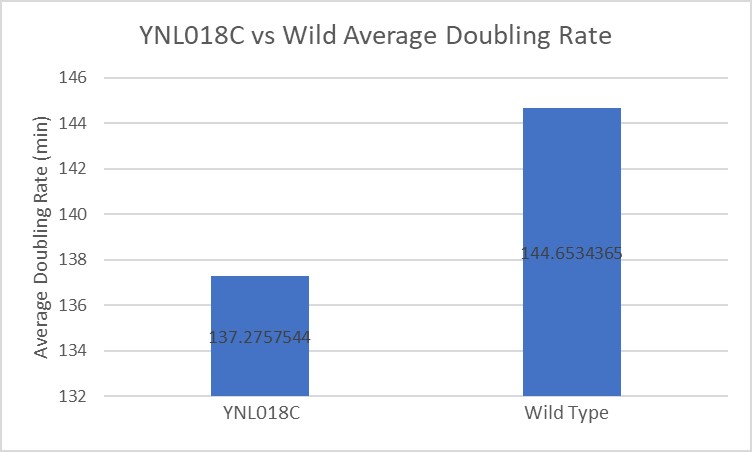
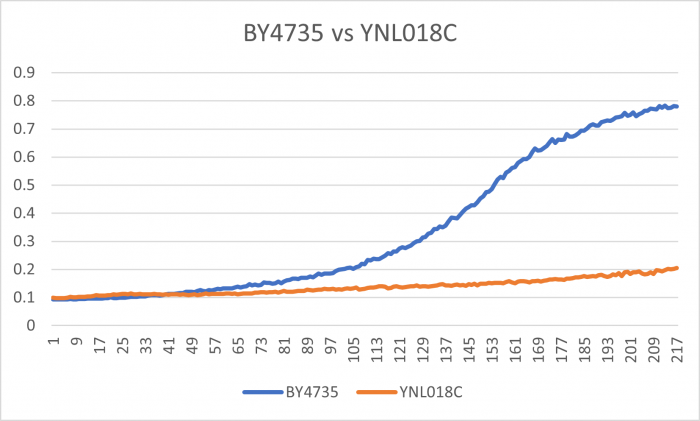
| − | Under normal conditions, the | + | Under normal conditions, the BY4735 doubling time is 144 minutes, while that of YNL018C is 145 minutes. This proves that both of these strains grow at a similar rate. When grown in an environment with hydroxychloroquine, YNL018C doubled in 211 minutes, while BY4735 doubled in 276 minutes. Both strains are inhibited by hydroxychloroquine, but BY4735 growth is staggered more. In hydroxychloroquine, BY4735 has a longer lag phase than that of YNL018C. (These doubling times and curves are the means of three experiments.) |
===[[UW-Stout/PMSF SP21|Phenylmethylsulphonyl Fluoride]]=== | ===[[UW-Stout/PMSF SP21|Phenylmethylsulphonyl Fluoride]]=== | ||
Revision as of 19:06, 28 April 2021
Share your knowledge...Edit this entry! <protect>
| Systematic name | YNL018C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr XIV:601774..599936 |
| Primary SGDID | S000004963 |
Description of YNL018C: Putative protein of unknown function[1]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
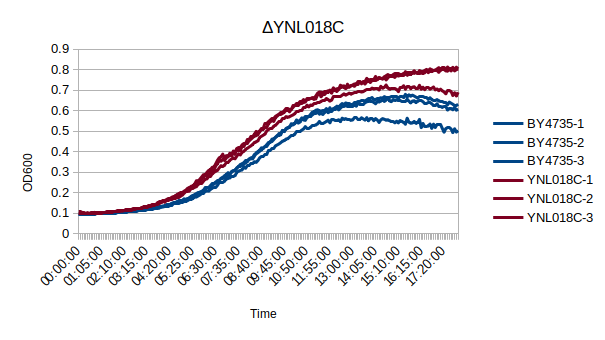
Growth Curve
In a BY4735 background, knocking out YNL018C seems to have a modest effect on growth rate, if any. In this assay, the BY4735 strain's doubling time was 162 minutes, while the YNL018C knock-out strain's doubling time was 129 minutes. (These doubling times are the means of three experiments.)
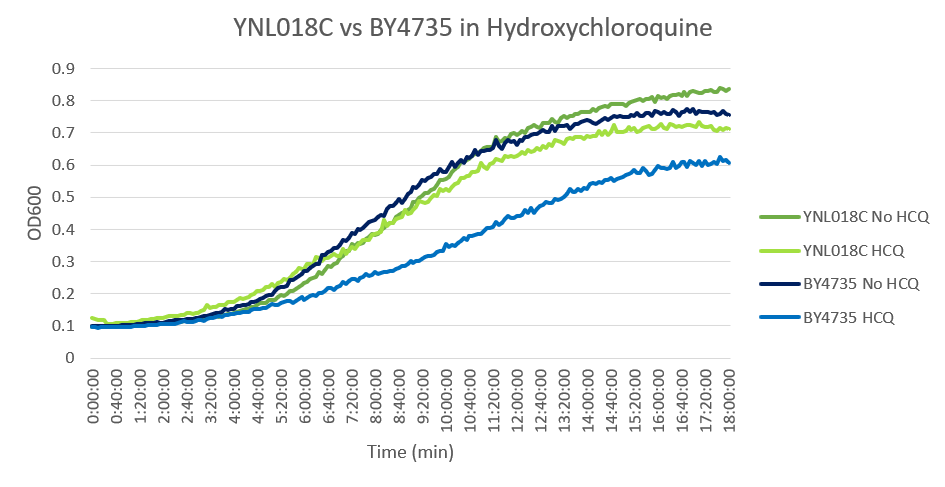
Hydroxychloroquine
HCQ refers to hydroxychloroquine
Under normal conditions, the BY4735 doubling time is 144 minutes, while that of YNL018C is 145 minutes. This proves that both of these strains grow at a similar rate. When grown in an environment with hydroxychloroquine, YNL018C doubled in 211 minutes, while BY4735 doubled in 276 minutes. Both strains are inhibited by hydroxychloroquine, but BY4735 growth is staggered more. In hydroxychloroquine, BY4735 has a longer lag phase than that of YNL018C. (These doubling times and curves are the means of three experiments.)
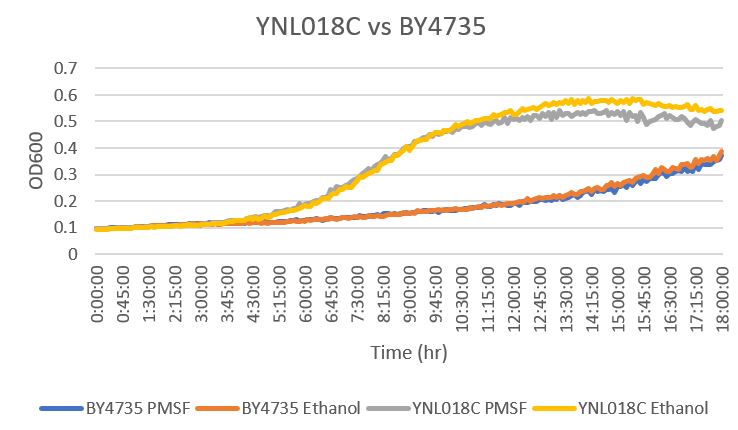
Phenylmethylsulphonyl Fluoride
The YNL018C yeast gene was determined to have an average doubling time of 151 minutes under the stress of the drug PMSF. This is is nearly 3 times as fast as the BY4735, which had a doubling time of 382 minutes. This was determined to be likely due to the modified yeast strain having a stronger resistance to the drug, making it able to grow faster and with less of an effect. Without the PMSF, the corresponding doubling times were 204 minutes for the YNL018C strain, and 442 minutes for the BY4735 strain.
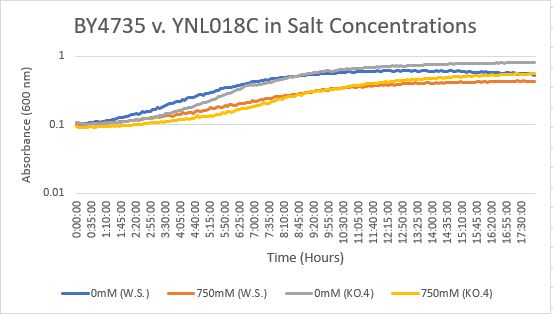
Salt Concentration (NaCl)
0mM NaCl conc. BY4735 strain's doubling time: 169 minutes
0mM NaCl conc. YNL018C strain's doubling time: 128 minutes
750mM NaCl conc. BY4735 strain's doubling time: 294 minutes
750mM NaCl conc. YNL018C strain's doubling time: 195 minutes
The graph above shows the growth rate for the previously listed strains and the relative level of NaCl concentration. Knocking out the gene positively influences the growth rate. Comparing the YNL018C doubling times between 0mM and 750mM NaCl, there is a moderate effect on the growth rate. This effect negatively influences the knockout strain's (YNL018C) growth rate.
Caffeine Group 1
Adding caffeine to the YNL018C strain had a negative on the yeast's growth rate. The doubling times are as followed:
Doubling time of YNL018C with caffeine: 131 minutes
Doubling time of YNL018C without caffeine: 295 minutes
Doubling time of BY4735 with caffeine: 138 minutes
Doubling time of BY4735 without caffeine: 187 minutes
Caffeine effected this strain by stunting its growth rate.
Competative Co-culture
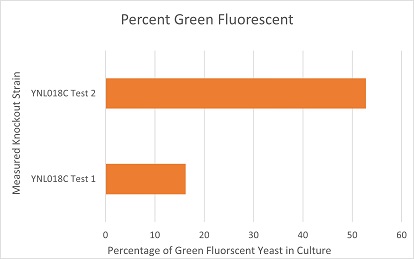
These are the results of a competitive co-culture protocol. The knockout strain was grown in a culture to test fitness against a green, fluorescent wild-type strain. The percentage of analyzed cells in the culture measured was those displaying green fluorescence. In theory, this should mean if a knockout has reduced the fitness of a strain of yeast, the fluorescent wild-type strain would have a higher percentage than the knockout strain. If the knockout does not decrease fitness, they would be roughly equal. This may not be so in results. Sources of error may include contamination and human error.
The results here are not expected in theory. Test 1 appears to have been contaminated with an organism that grows faster than yeast and does not display a fluorescent trait like the wild-type. Test 2 yields an expected result that suggests the knockout of YNL018C does not reduce fitness in a fair, competitive co-culture.
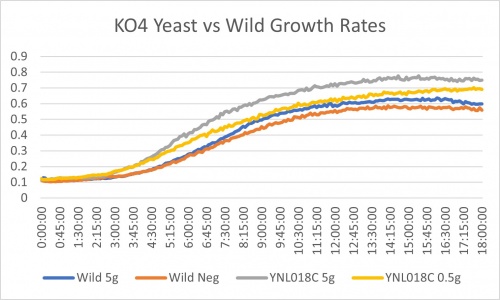
Ammonium Sulfate
YNL018C gene shows an interesting result. While the wild strain from 5g/ml of ammonium sulfate to none shows a 17.8% slower rate, it shows a 40.8% slower rate. For further analysis 0.5g/ml to no ammonium sulfate was compared too. Wild showed 1% slower rate and YNL018C showed 31.4% slowed rate. Based on the differences of these two treatments in the wild to the YNL018C KO strain it could be said that there was experimental error. The differences in percent are 16.8% in the wild and roughly 8.6% in the KO strain. The doubling time of the negative control is like the wild as well. These two points could indicate some form of experimental error. It could also be that this strain with the knocked out YNL018C gene is in fact more sensitive to nitrogen starvation
<protect>
UV Light
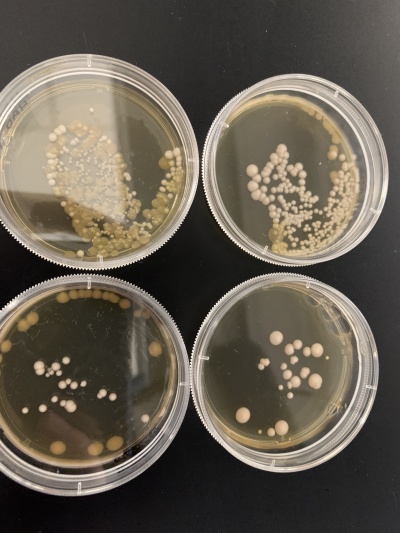
Results:
- Experiment 1:0sec=234 colonies, 600sec=25 colonies.
- Experiment 2:0sec=136 colonies, 600sec=22 colonies.
Interpretation: The two 0sec plates are on top and the 600sec plates are on the bottom. There appears to be some contamination on the 0sec experiment 1 plate (top right) and a lot of contamination on the experiment 2 plates (left). The yeast cells seemed to be significantly affected by the UV light with about 80-90% of the cells being killed compared to the control. It's possible that the knocked-out gene was related to UV light exposure.
Calcofluor White
From this data we can determine that there was minimal growth inhibition on this strain of yeast. The modified yeast had a doubling rate that was ten minutes faster than the wild type on average, and when compared to the effect on other yeast strains this indicates there was no growth slowing effect on the modified yeast.
Heat Shock
The top plate is the non-heat shocked sample, the middle and bottom are heat shocked. The results from this experiment showed that the YNLo19C strain of yeast is not heat sensitive. All three of the samples have similar growth and are therefore not heat sensitive.
Caffeine Group 2
References
See Help:References on how to add references
See Help:Categories on how to add the wiki page for this gene to a Category </protect>