Difference between revisions of "SGD Newsletter, Spring 2019"
(→25th Anniversary of the SGD Website) |
(→SGD Newsletter, Spring 2019) |
||
| Line 2: | Line 2: | ||
=SGD Newsletter, Spring 2019= | =SGD Newsletter, Spring 2019= | ||
'''About this newsletter:'''<br> | '''About this newsletter:'''<br> | ||
| − | This is the Spring 2019 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. | + | This is the Spring 2019 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community.[[File:cdc6.png|thumb|right|upright=.8|link=https://wiki.yeastgenome.org/images/f/f8/cdc6.png|The CDC6 locus page from 1994 (left) and the [https://www.yeastgenome.org/locus/S000003730 current CDC6 page]]] |
| + | |||
__TOC__ | __TOC__ | ||
| Line 17: | Line 18: | ||
SGD has now incorporated proteome-wide protein abundance data obtained from a comprehensive meta-analysis by Ho et al., 2018. The authors normalized and combined 21 different S. cerevisiae protein abundance datasets—including data from both untreated cells and cells treated with various environmental stressors—to create a unified protein abundance dataset where all values are in the intuitive units of molecules per cell. | SGD has now incorporated proteome-wide protein abundance data obtained from a comprehensive meta-analysis by Ho et al., 2018. The authors normalized and combined 21 different S. cerevisiae protein abundance datasets—including data from both untreated cells and cells treated with various environmental stressors—to create a unified protein abundance dataset where all values are in the intuitive units of molecules per cell. | ||
| − | Normalized abundance measurements and associated metadata from untreated and treated cells are displayed in tabular form in the experimental data section of protein-tabbed pages (e.g. CDC28). Several different controlled vocabularies have been employed to standardize the metadata display. In addition, calculated median abundance and median absolute deviation (MAD) values are displayed in the protein section of Locus Summary pages (e.g. PHO85). Two new [https://yeastmine.yeastgenome.org/yeastmine/begin.do YeastMine] templates have been created to provide access to these data: [https://yeastmine.yeastgenome.org/yeastmine/template.do?name=Gene_ProteinAbundance&scope=all Gene → Protein Abundance] and [https://yeastmine.yeastgenome.org/yeastmine/template.do?name=Protein_Median_Abundance&scope=all Gene → Median Protein Abundance]. | + | Normalized abundance measurements and associated metadata from untreated and treated cells are displayed in tabular form in the experimental data section of protein-tabbed pages (e.g. CDC28). Several different controlled vocabularies have been employed to standardize the metadata display. In addition, calculated median abundance and median absolute deviation (MAD) values are displayed in the protein section of Locus Summary pages (e.g. PHO85). |
| + | |||
| + | Two new [https://yeastmine.yeastgenome.org/yeastmine/begin.do YeastMine] templates have been created to provide access to these data: [https://yeastmine.yeastgenome.org/yeastmine/template.do?name=Gene_ProteinAbundance&scope=all Gene → Protein Abundance] and [https://yeastmine.yeastgenome.org/yeastmine/template.do?name=Protein_Median_Abundance&scope=all Gene → Median Protein Abundance]. | ||
== Alliance of Genome Resources: 2.1 Release== | == Alliance of Genome Resources: 2.1 Release== | ||
Revision as of 12:50, 10 May 2019
SGD Newsletter, Spring 2019
About this newsletter:
This is the Spring 2019 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community.
Contents
- 1 SGD Newsletter, Spring 2019
25th Anniversary of the SGD Website
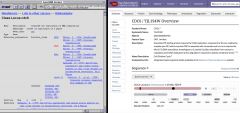
Wednesday, May 8th, marked the 25th year that the SGD website has been live. SGD staff celebrated the day by taking a tour around the Stanford campus and enjoying the beautiful California spring weather. We went up and saw some fantastic views from the 14th floor of Hoover Tower, then hiked to the top of the Stanford Dish where we could even see the San Francisco skyline. Of course, we also had cake after that! Did you catch our throwback page for CDC6?
S288C Transcriptome and New Data Tracks in JBrowse
We have recently equipped our genome browsing tool JBrowse with 9 new Transcriptome data tracks, making JBrowse an even more powerful way to explore the vast heterogeneity of the S288C transcriptome. These information-rich data tracks visualize RNA transcripts from the TIF-seq dataset published by Pelechano et al. (2013), enabling quick and easy viewing of the position, length, and abundance of transcript isoforms sequenced in the study.
SGD has also updated our JBrowse with an additional 157 new data tracks related to genome-wide experiments and omics data for you to explore. The categories added include: Transcription & Transcriptional Regulation; Histone Modification; Chromatin Organization; RNA Catabolism; Transposons; DNA Replication, Recombination, and Repair.
Proteome-wide Abundance Data
SGD has now incorporated proteome-wide protein abundance data obtained from a comprehensive meta-analysis by Ho et al., 2018. The authors normalized and combined 21 different S. cerevisiae protein abundance datasets—including data from both untreated cells and cells treated with various environmental stressors—to create a unified protein abundance dataset where all values are in the intuitive units of molecules per cell. Normalized abundance measurements and associated metadata from untreated and treated cells are displayed in tabular form in the experimental data section of protein-tabbed pages (e.g. CDC28). Several different controlled vocabularies have been employed to standardize the metadata display. In addition, calculated median abundance and median absolute deviation (MAD) values are displayed in the protein section of Locus Summary pages (e.g. PHO85).
Two new YeastMine templates have been created to provide access to these data: Gene → Protein Abundance and Gene → Median Protein Abundance.
Alliance of Genome Resources: 2.1 Release
In March, the Alliance released version 2.1. The release showcases the combined effort from SGD and the other core Alliance members. Notable improvements and new features include:
- Disease Associations file on Downloads page
- Gene Descriptions files on Downloads page
- Updated Interactions table, including link outs to MOD pages
- Updated Gene Ontology and Expression ribbon displays
- Related Data links in Search results
SGD at Biocuration 2019
From April 7th-10th, SGD attended the International Society for Biocuration's 12th International Biocuration Conference in Cambridge, UK. Several of our staff presented posters, while Senior Biocuration Scientist Edith Wong gave a great talk on her recent Database publication: Integration of Macromolecular Complex Data into the Saccharomyces Genome Database. Below are the posters and the talk SGD staff presented at Biocuration 2019. Click on any of the links below to download the poster.
Presentation
| Presenter | Presentation Title |
| Edith Wong | "Integration of Macromolecular Complex Data into the Saccharomyces Genome Database" |
Posters
| Presenter | Poster Title |
| Suzi Aleksander | "In the Know About GO: A Newly Redesigned Website for the Gene Ontology" |
| Felix Gondwe | "Downloading Data from SGD" |
| Edith Wong | "Integration of Macromolecular Complex Data into the Saccharomyces Genome Database" |
Recent publications from SGD Staff
- Wong ED, Skrzypek MS, Weng S, Binkley G, Meldal BHM, Perfetto L, Orchard SE, Engel SR, Cherry JM; SGD Project (2018). Integration of Macromolecular Complex Data into the Saccharomyces Genome Database. Database (Oxford). 2019 Jan 1; 2019. doi: 10.1093/database/baz008 PMID:30715277
SGD Biocurators Out and About
- Biocuration Scientist Kevin MacPherson gave tutorials at the Fungal Pathogen Genomics course at the Wellcome Genome Campus in May. He taught students how to use tools and resources at both SGD and the Candida Genome Database.
- Several SGD representatives attended the 2019 Gene Ontology Consortium Meeting in Cambridge, UK, in early April.
- In March, Biocuration Scientist Kevin MacPherson and Senior Biocuration Scientist Barbara Dunn attended the Genetics Society of America's 30th Fungal Genetics Conference in Pacific Grove, CA. They both presented posters.
| Presenter | Poster Title |
| Barbara Dunn | "Associating Yeast Genes with Human Disease-related Genes at SGD" |
| Kevin MacPherson | "Comparative Genomics at the Saccharomyces Genome Database" |
Upcoming Meetings
14th Yeast Lipid Conference - YLC 2019
Ljubljana, Slovenia
May 22 to May 24, 2019
Yeast ORFan Gene Project: Finding a place for ORFans to GO
Austin College, Sherman, TX
June 17 to June 21, 2019
7th Conference on Physiology of Yeasts & Filamentous Fungi (PYFF)
Milan, Italy
June 24 to June 27, 2019
6th International Synthetic & Systems Biology Summer School - SSBSS 2019
Scuola Normale Superiore, Pisa, Tuscany, Italy
July 22 to July 26, 2019
CSHL Yeast Genetics & Genomics course
Cold Spring Harbor, NY
July 23 to August 12, 2019
29th International Conference on Yeast Genetics and Molecular Biology (ICYGMB)
Gothenburg, Sweden
August 18 to August 22, 2019
35th International Specialised Symposium on Yeasts (ISSY)
Antalya, Turkey
October 21 to October 25, 2019
Yeast Research: Origins, Insights, Breakthroughs
Cold Spring Harbor, NY
October 23 to October 26, 2019
The Allied Genetics Conference - TAGC 2020
Metro Washington, DC
April 22 to April 26, 2020

