Difference between revisions of "YGL140C"
(→Caffeine) |
|||
| (4 intermediate revisions by the same user not shown) | |||
| Line 53: | Line 53: | ||
The protocol can be found at | The protocol can be found at | ||
| + | |||
===[[UW_Stout/UV_Light_FA19|UV Light Exposure]]=== | ===[[UW_Stout/UV_Light_FA19|UV Light Exposure]]=== | ||
| Line 96: | Line 97: | ||
The Wild Type BY4735 has data located at the Nitrogen Starvation Protocol. | The Wild Type BY4735 has data located at the Nitrogen Starvation Protocol. | ||
==[[UW-Stout/Nitrogen_starvation_FA19|Nitrogen starvation]]== | ==[[UW-Stout/Nitrogen_starvation_FA19|Nitrogen starvation]]== | ||
| + | |||
| + | == [https://wiki.yeastgenome.org/index.php?title=UW-Stout/Hydroxyurea_FA19&oldid=401696 Hydroxyurea] == | ||
| + | [[File:AvaYGL140C.png]] | ||
| + | The averaging doubling time for YGL140C is 194.83 minutes. The averaging doubling time for the unstressed YGL140C is 352.74 minutes | ||
<!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | <!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | ||
| Line 107: | Line 112: | ||
[[File:JJ10.PNG]] | [[File:JJ10.PNG]] | ||
[[File:JaredJ10.PNG]] | [[File:JaredJ10.PNG]] | ||
| + | |||
| + | The bar graph represents the zone of inhibition of the knockout strain versus the wild type. Our data collected shows that the knockout strain saw a larger zone of inhibition (cells were not able to grow there). The error bar on the knockout strain represents the standard deviation of the experiment and how recreatable our results were with these results. Using these observations it is evident that the yeast strain YGL140C undergoes more stress from the oxidative properties of 30 percent hydrogen peroxide. The zone of inhibition is larger because the yeast cells are not able to grow as well as the wild type can in these conditions. For the strain YGL140C our plate values were as follows (in cm) 2.35, 2.3 and 2.4 compared to our wild type plate which stayed constant 2.1, 2.1 and 2.2 cm | ||
<protect> | <protect> | ||
Latest revision as of 14:39, 5 May 2022
Share your knowledge...Edit this entry! <protect>
| Systematic name | YGL140C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr VII:245012..241353 |
| Primary SGDID | S000003108 |
Description of YGL140C: Putative protein of unknown function; non-essential gene; contains multiple predicted transmembrane domains[1][2][3]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
This gene is part of the UW-Stout Orphan Gene Project. Learn more here.
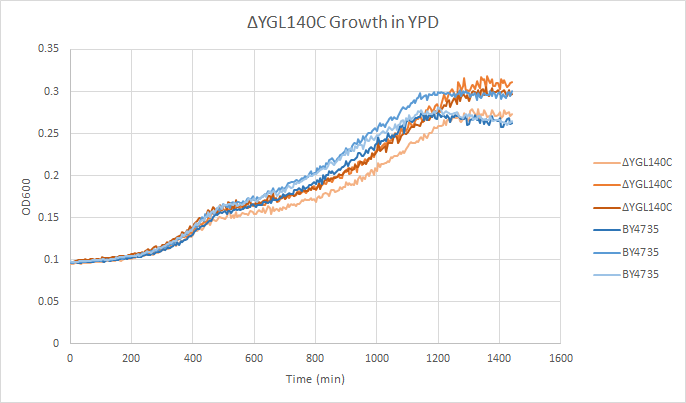
Growth Curve
In a BY4735 background, knocking out YGL140C seems to have little or no effect on growth rate in log-phase. In this assay, the BY4735 strain's doubling time was 414 minutes, while the YGL140C knock-out strain's doubling time was 432 minutes. (These doubling times are the means of three experiments.)
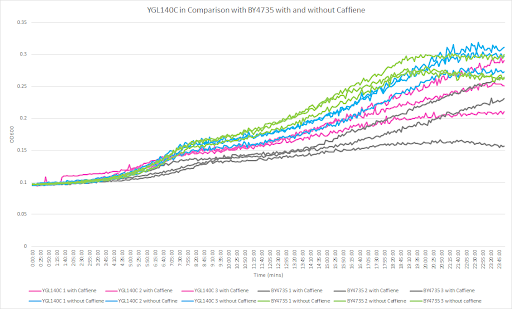
Caffeine and Yeast Cells
This graph shows the growth curve with and without caffeine. It has the knockout strain and wild type yeast cells. The caffeine enhanced the growth in both wild type and knockout strain. The average doubling time between the three experiments for the knockout strand was 777.97 minutes. The caffeine also had a positive effect on the growth curve for the wild type with the average doubling time being 1245.02 minutes. The results were concluded from the wild type and knockout strand was with this amount of caffeine it leads to an increase in growth.
The protocol can be found at
Caffeine
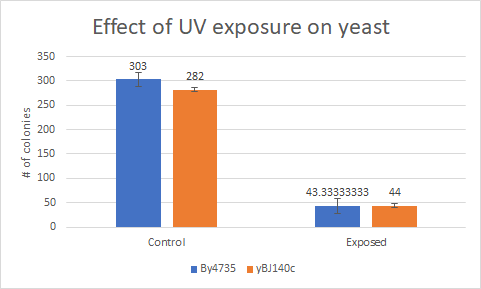
This bar graph shows that compared to a BY4735 background' Knocking out yGL140c have no effect on how sensitive yeast is to UV Light
The protocol can be found at
UV Light Exposure
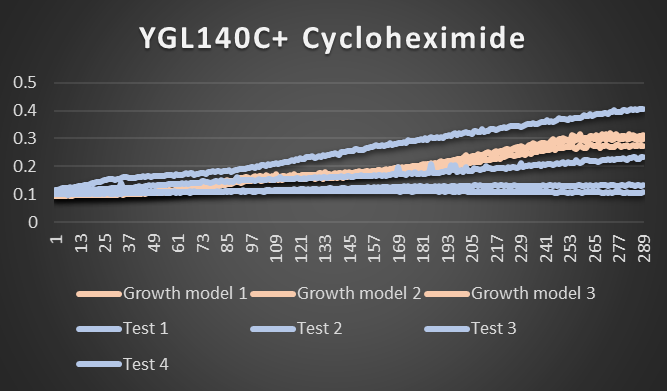
Cycloheximide effecting YGL140C
As you can see from the picture above test 4 did really well and survived while tests 1-3 were disabled and didn't function as well. Growth models 1-3 are the factors that were not stressed.
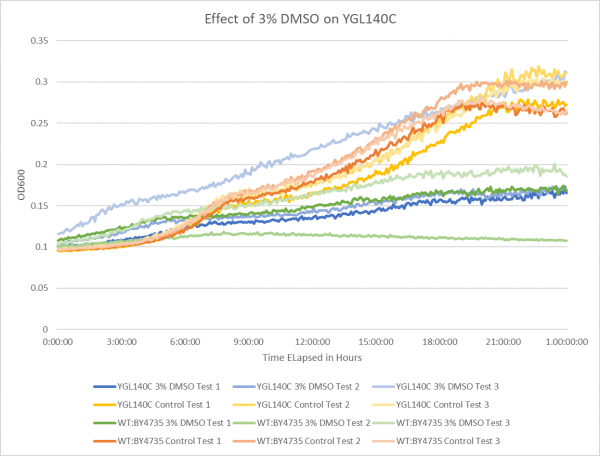
Effect of 3% DMSO on YGL140C
The graph above shows the effect of 3% DMSO on YGL140C. While examining the data from test 3 it is thought that the 3% DMSO test on YGL140C was contaminated; the results are still shown on the graph but the information was not used during growth rate calculations. From the data represented the average doubling rate for the YGL140C knockout strain without DMSO was 561 minutes. When DMSO is added the average doubling time increased to 1118 minutes. This shows that 3% DMSO did add stress to this knockout strain. The average doubling time of WT:BY4735 without DMSO was 514 minutes. The average doubling rate for WT:BY4735 with DMSO was 1529 minutes. If we compare the average doubling rate of WT:BY4735 with DMSO to YGL140C with DMSO we can see that without the YGL140C gene the yeast was able to grow more quickly under DMSO induced stress. It is also observed that both control groups had a very close growth rate, which means that the YGL140C gene has a minimal effect of yeast growth in optimal conditions.
Growth in Proline and Ammonium Sulfate
The YGL140C strand grew faster in the proline media with an average doubling time of 34.43 minutes. It was not that much faster than growing in ammonium sulfate media though because that media was only two minutes slower with an average doubling time of 36.73 minutes. In the wild type strand though, the wild type grew faster in the ammonium sulfate media with an average doubling time of 41.53 minutes compared to the 47.27 minutes it took in proline media.
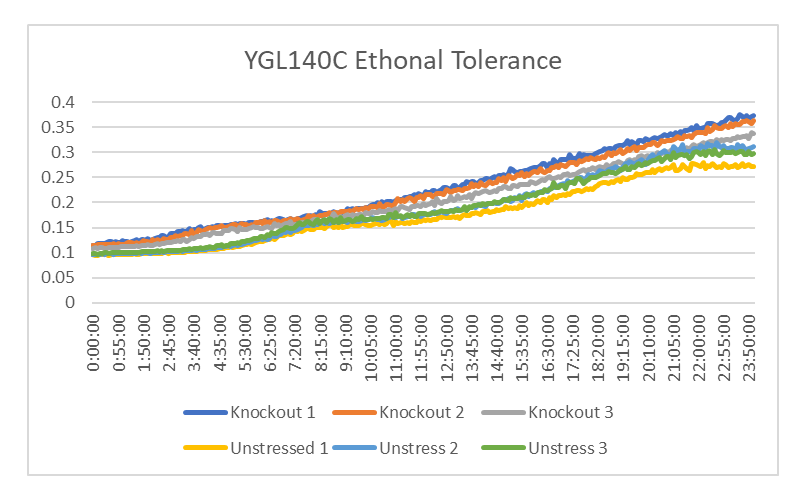
Ethanol Tolerance in YGL140C
This graph shows the rate of growth between YGL140C treated and untreated with ethanol. The rate of growth of the yeast treated with ethanol is 818.32 minutes. The rate of growth of the yeast untreated with ethanol is 1903.21 minutes. This shows that the strands of YGL140C treated with ethanol had a decrease rate of growth compared to those who weren't.
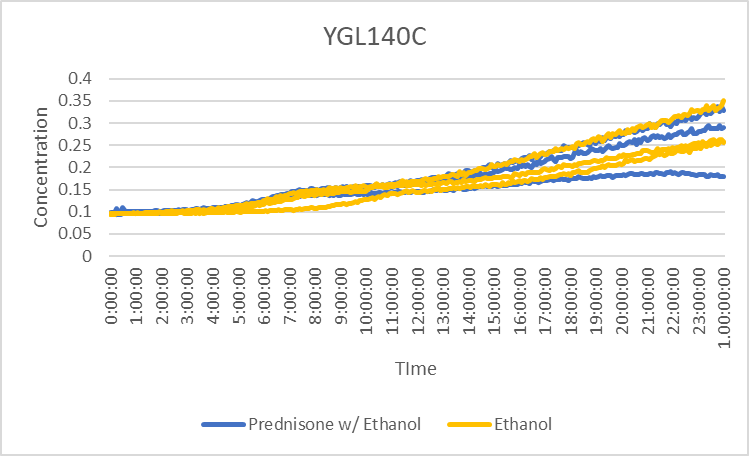
Prednisone Effects on YGL140C
The graph above shows how dilutions P2 (30 µg/ml) and E2 (10% ethanol solution) impacted the growth rate of the knockout yeast strain YGL140C over 24 hours. The concentration did not double for test 3 of P2 for this experiment, so by taking only the first two test results, the doubling rate averages to around 990 minutes for only ethanol. As for the Prednisone with ethanol, the doubling rate averages to around 900 minutes. The ethanol decreased the rate of growth by almost 90 minutes. These results conclude that Prednisone promotes the growth of yeast cells without the gene YGL140C.
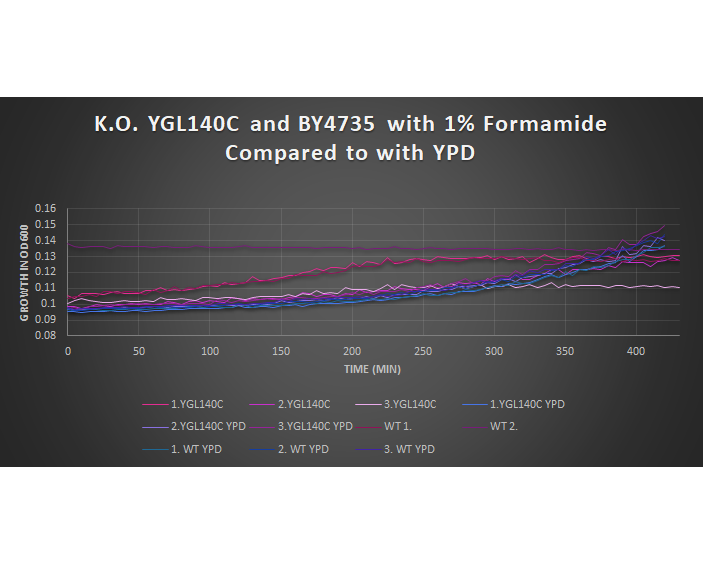
1% Formamide Yeast Torture[[1]]
This graph shows the growth curve of the knockout yeast strain YGL140C and wild type BY4735 in 1% formamide in 3 trials. The average calculated doubling time for YGL140C with 1% formamide was 3,420.64 minutes, while the unstressed strain was 1,988.91 minutes. This concluded that the stressed out strain is sensitive to 1% formamide, and grows at a slower pace in this environment. Our stressed wild types doubling time was 2,865.53 minutes, and the unstressed was 2,230.85 minutes. The unstressed wild type in this experiment proved to show better growth concluding that the addition of 1% formamide for this strain does cause sensitivity.
The Wild Type BY4735 has data located at the Nitrogen Starvation Protocol.
Nitrogen starvation
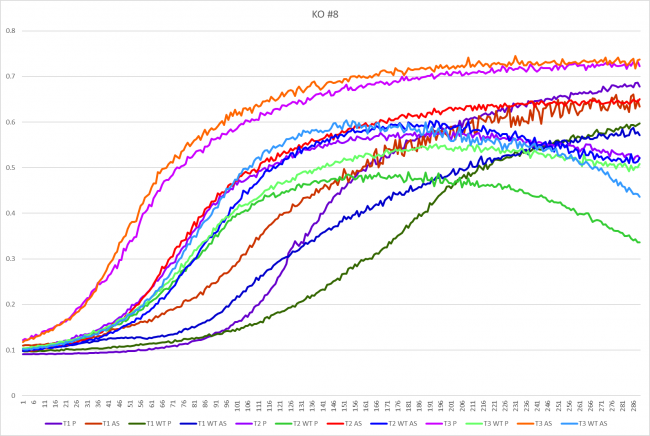
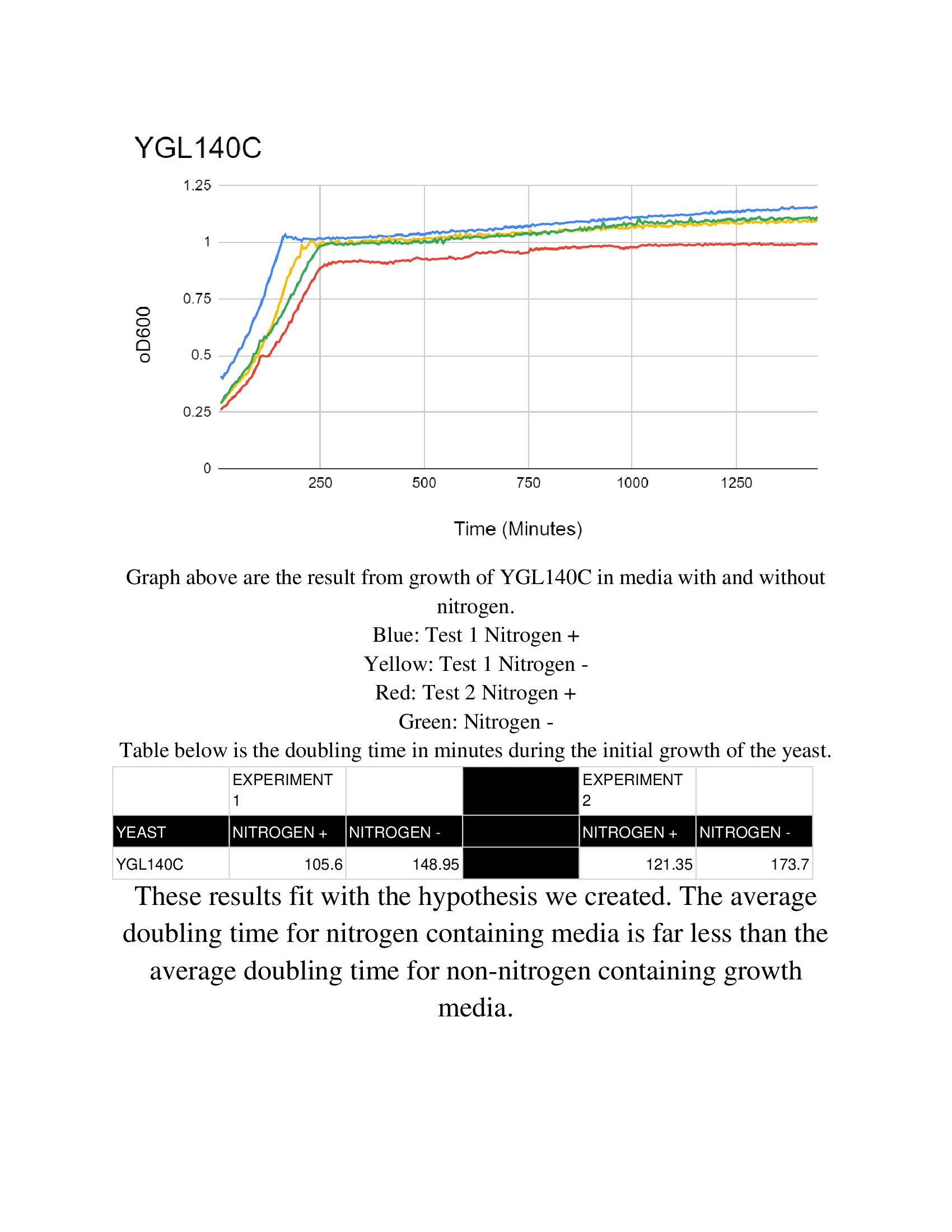
Hydroxyurea
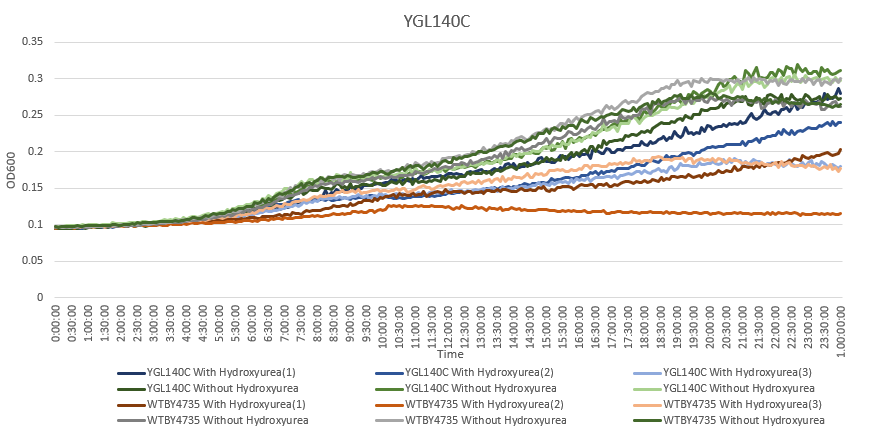 The averaging doubling time for YGL140C is 194.83 minutes. The averaging doubling time for the unstressed YGL140C is 352.74 minutes
The averaging doubling time for YGL140C is 194.83 minutes. The averaging doubling time for the unstressed YGL140C is 352.74 minutes
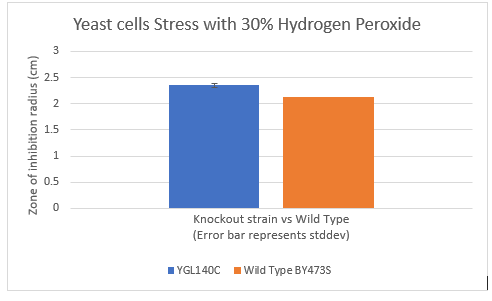
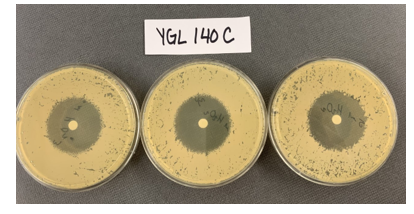
Zone of Inhibition of 30% Hydrogen Peroxide on YGL140C
The bar graph represents the zone of inhibition of the knockout strain versus the wild type. Our data collected shows that the knockout strain saw a larger zone of inhibition (cells were not able to grow there). The error bar on the knockout strain represents the standard deviation of the experiment and how recreatable our results were with these results. Using these observations it is evident that the yeast strain YGL140C undergoes more stress from the oxidative properties of 30 percent hydrogen peroxide. The zone of inhibition is larger because the yeast cells are not able to grow as well as the wild type can in these conditions. For the strain YGL140C our plate values were as follows (in cm) 2.35, 2.3 and 2.4 compared to our wild type plate which stayed constant 2.1, 2.1 and 2.2 cm <protect>
References
See Help:References on how to add references
- ↑ Giaever G, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418(6896):387-91 SGD PMID 12140549
- ↑ Voet M, et al. (1997) The sequence of a nearly unclonable 22.8 kb segment on the left arm chromosome VII from Saccharomyces cerevisiae reveals ARO2, RPL9A, TIP1, MRF1 genes and six new open reading frames. Yeast 13(2):177-82 SGD PMID 9046099
- ↑ Volckaert G, et al. (2003) Disruption of 12 ORFs located on chromosomes IV, VII and XIV of Saccharomyces cerevisiae reveals two essential genes. Yeast 20(1):79-88 SGD PMID 12489128
See Help:Categories on how to add the wiki page for this gene to a Category </protect>