Difference between revisions of "YMR144W"
(→Heat Shock) |
(→Heat Shock) |
||
| Line 37: | Line 37: | ||
==Heat Shock== | ==Heat Shock== | ||
{| class="wikitable" | {| class="wikitable" | ||
| − | |+ | + | |+ YMR144W Yeast Colonies at 50 degrees Celsius |
|- | |- | ||
! Time (sec) !! Colonies !! | ! Time (sec) !! Colonies !! | ||
Revision as of 11:15, 2 May 2023
Share your knowledge...Edit this entry! <protect>
| Systematic name | YMR144W |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr XIII:553362..554390 |
| Primary SGDID | S000004752 |
Description of YMR144W: Putative protein of unknown function; localized to the nucleus; YMR144W is not an essential gene[1][2]
</protect>
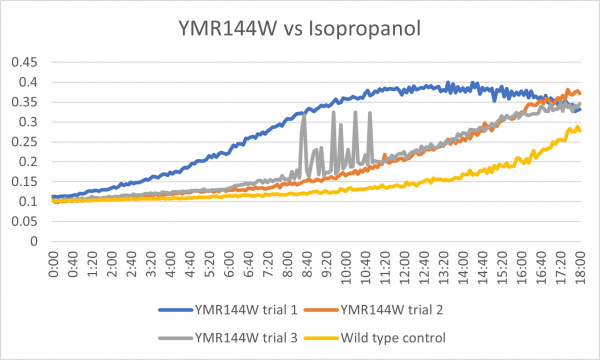
Isopropanol
Analysis
Trial two was the most linear to our wild type control while trials one and three seem to be inconclusive.
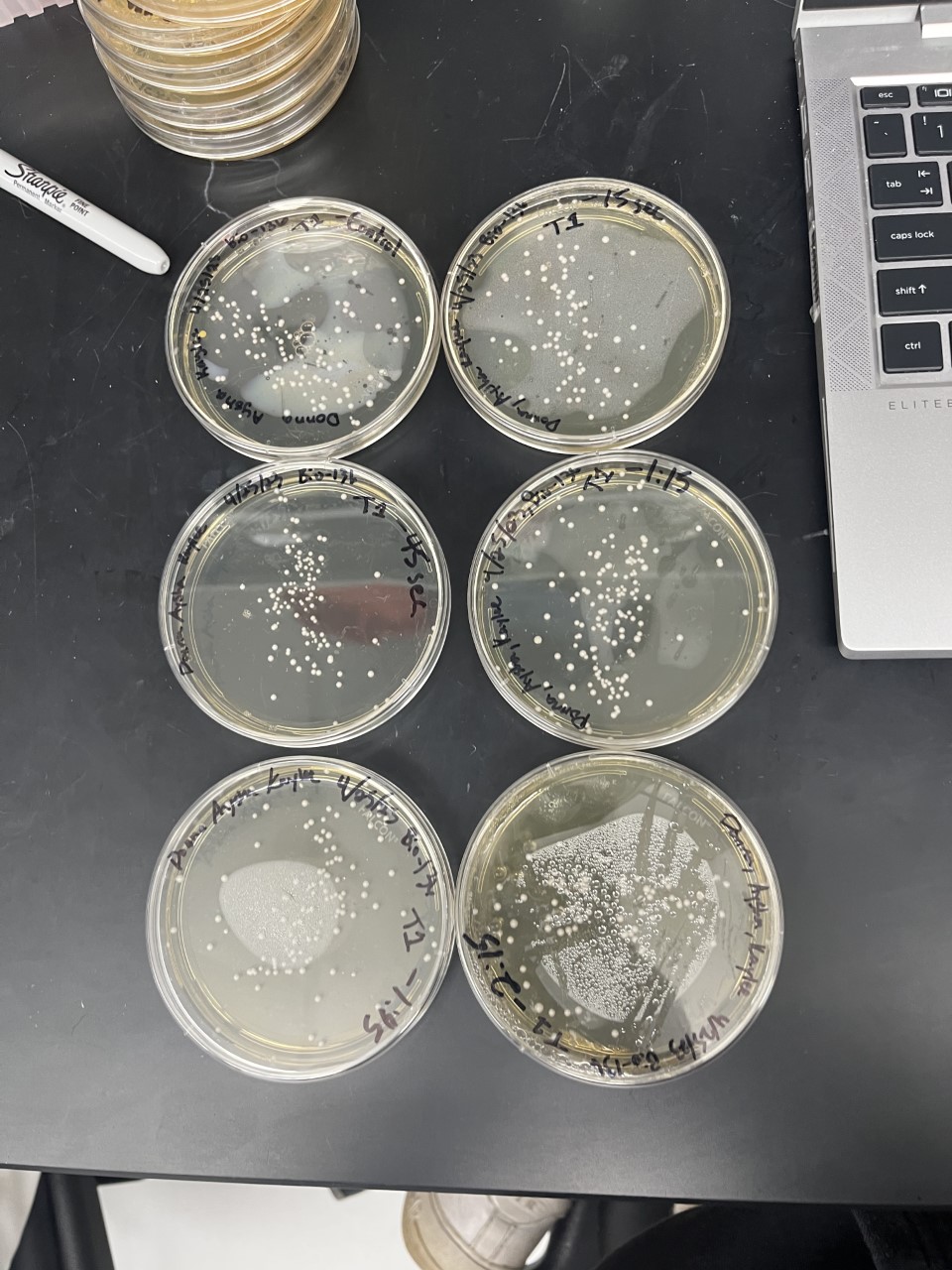
Heat Shock
| Time (sec) | Colonies | |
|---|---|---|
| 0 | >100 | |
| 15 | 92 | |
| 45 | 86 | |
| 75 | 100 | |
| 105 | 84 | |
| 135 | 81 |
The general trend of our data yielded a negative relationship between time of heat shock. It can be inferred that with a longer amount of time we the yeast cells could continue being killed off until all of them died.
Bases
<protect>
References
See Help:References on how to add references
See Help:Categories on how to add the wiki page for this gene to a Category </protect>