UW-Stout/PMSF SP21
Contents
Materials
- Corning COSTAR 96-well clear flat-bottom assay plate
- Wild-type yeast in 2x synthetic complete media at an OD600 of 0.1-0.2
- Phenylmethylsulphonyl fluoride, 100 mM in ethanol
- Ethanol
- P20, P200, P1000 micropipettors
Equipment
- Molecular Devices SpectraMax Plus 384 Microplate Reader
Precautions
Use gloves, fume hood, and goggles, as PMSF is extremely hazardous. This chemical is known to be extremely corrosive and damaging to tissue and can house irreversible eye damage. Can form a dangerous gas upon contact with water, form a corrosive and toxic gas. If it comes in contact with metal it can form a flammable gas. For this reason, the PMSF will be dissolved in an ethanol solution.
Calibration Protocol
- Vortex the yeast culture briefly to resuspend the yeast cells.
- Prepare a stock solution of PMSF in ethanol solution of 0.10mM
- Create 5 different molarities of 0.05mM, 0.01mM, 0.005mM, 0.002mM, and 0.001mM
- In each well, place 1ul of each molarity, filling a total of 6 plates, each with a different molarity
- Set up 8 wells as follows:
- Well 1: 50ul of sterile water (Control with just sterile water)
- Well 2: 1uL of ethanol, 49uL of sterile water (Control with just ethanol and sterile water)
- Well 3: 1uL of 0.10mM, 49uL of sterile water
- Well 4: 1uL of 0.05mM, 49uL of sterile water
- Well 5: 1uL of 0.01mM, 49uL of sterile water
- Well 6: 1uL of 0.005mM, 49uL of sterile water
- Well 7: 1uL of 0.002mM, 49uL of sterile water
- Well 8: 1uL of 0.001mM, 49uL of sterile water
- Add 50uL of yeast culture to each well
- To measure cell growth over a period of 24 hours, set up the plate reader as follows:
- Temperature: 30°C
- Mode: Kinetic
- Wavelength: 600 nm
- Interval: 5 minutes
- Total run time: 24 hours
- Shake before reading: 30 seconds.
- Transfer the assay plate to the reader and read for 24 hours
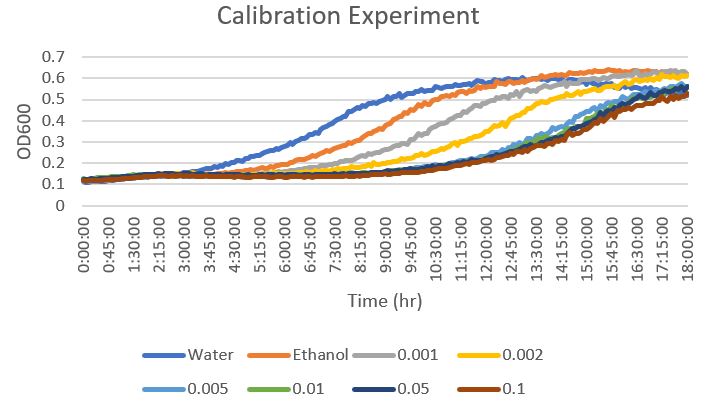
Raw Data From Calibration Experiment
From this data, a concentration of 0.002mM was determined to be an effective concentration to torture the yeast with phenylmethylsulphonyl fluoride. This is because it was not too detrimental on the growth of the yeast without allowing it to flourish.
Protocol
The data collected from the calibration protocol listed above has determined that a concentration of 0.002mM is an effective concentration for the torture of the yeast in this experiment
- Vortex the yeast culture briefly to resuspend the yeast cells
- Set up all 8 wells with 4 different yeast strains, with each yeast strain taking up 2 wells. To form a negative control, each strain must be exposed to just ethanol as well as the 0.002mM PMSF/Ethanol solution
- This needs to be done because the effects of just ethanol need to be known to understand the difference that the PMSF will have on each of the different yeast strains
- If more than 4 strains need to be testing, more wells will be needed. I accounted for this by dividing the testing amongst different days, or borrowing some space from fellow students
- Set up the 8 wells as follows:
- Well 1: 1ul of ethanol, 49ul of sterile water, 50ul of yeast strain 1
- Well 2: 1ul of 0.002mM, 49ul of sterile water, 50ul of yeast strain 1
- Well 3: 1ul of ethanol, 49ul of sterile water, 50ul of yeast strain 2
- Well 4: 1ul of 0.002mM, 49ul of sterile water, 50ul of yeast strain 2
- Well 5: 1ul of ethanol, 49ul of sterile water, 50ul of yeast strain 3
- Well 6: 1ul of 0.002mM, 49ul of sterile water, 50ul of yeast strain 3
- Well 7: 1ul of ethanol, 49ul of sterile water, 50ul of yeast strain 4
- Well 8: 1ul of 0.002mM, 49ul of sterile water, 50ul of yeast strain 4
- To measure cell growth over a period of 24 hours, set up the plate reader as follows:
- Temperature: 30°C
- Mode: Kinetic
- Wavelength: 600 nm
- Interval: 5 minutes
- Total run time: 24 hours
- Shake before reading: 30 seconds
- Transfer the assay plate to the reader and read for 24 hours.
- Repeat the experiment more than once, as variation is likely