SGD Quarterly Newsletter, Fall 2014
About this newsletter:
This is the Fall 2014 issue of the quarterly SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can also subscribe to SGD's RSS feed to receive updates on SGD news: http://www.yeastgenome.org/feed
Contents
SGD User Survey
We are preparing for our grant renewal due to be submitted next spring. For a successful application we need your opinion and experience with SGD. Please fill out our survey here!
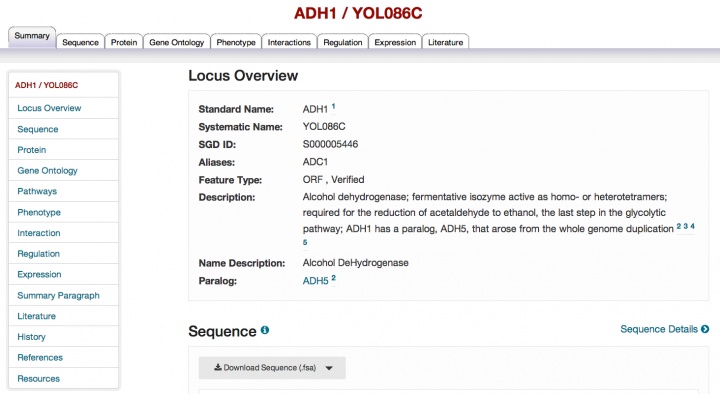
New and improved Locus Summary pages
We are pleased to announce that the redesign of our gene-specific pages, which has been ongoing over the past year, is now complete with the release of the reworked Locus Summary page. The page contains all of the information on the previous Locus Summary page and more, and has a more modern look and feel. Note that the order and organization of the sections has changed and the order of the tabs across the top of the page has changed as well. The new elements on the page include a navigation bar on the left to take you to the different sections of the page, a redesigned map showing genomic context in the sequence section, and a new interactive histogram summarizing expression data. Biochemical pathway information now appears in its own section (see an example) and we have added a History section to replace the previous Locus History tab. If there are no data of a particular type (for example, Pathways), then that section is absent from the page.
Please explore this new page and send us your feedback.
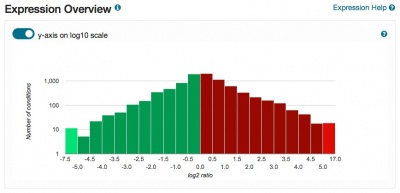
Redesigned Expression pages
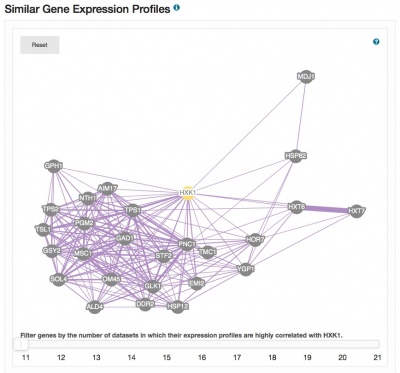
The Expression pages have been redesigned and now include a clickable histogram depicting conditions and datasets in which the gene of interest is up- or down-regulated. Expression data are derived from records contained in the Gene Expression Omnibus, and datasets are assigned one or more categories to facilitate grouping, filtering and browsing. Short descriptions of the focus of each experiment are also provided. The PCL files generated for each dataset are used to populate the expression analysis tool SPELL. Also included on the pages are network diagrams which display genes that share expression profiles. The Expression pages provide seamless access to the SPELL tool at SGD, as well as external resources such as Cyclebase, GermOnline, YMGV and FuncBase.
Please explore these new pages, accessible via the Expression tab on your favorite Locus Summary page, and send us your feedback.
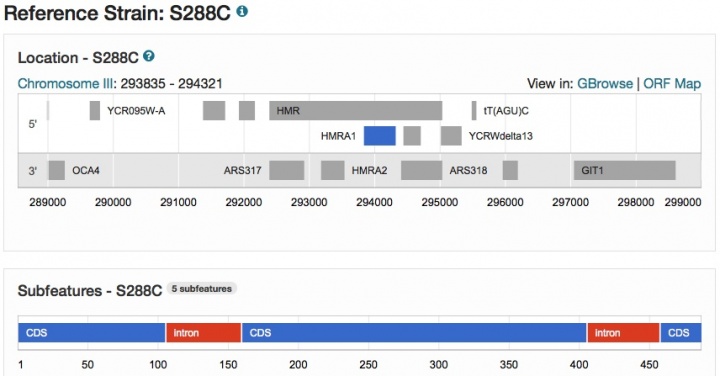
New Sequence, Chromosome, and Contig pages
New Sequence pages are now available in SGD for virtually every yeast gene (e.g., HMRA1 Sequence page), and include genomic sequence annotations for the Reference Strain S288C, as well as several Alternative Reference Genomes from strains such as CEN.PK, RM11-1a, Sigma1278b, and W303 (more Alternative References coming soon). Each page includes an Overview section containing descriptive information, maps depicting genomic context in Reference Strain S288C (as shown below) and Alternative Reference strains, as well as chromosomal and relative coordinates in S288C.
The sequence itself includes display options for genomic DNA, coding DNA, or translated protein.
Also available on each Sequence page are links to redesigned S288C Chromosome pages, links to new Contig pages for Alternative Reference Genomes, and a Downloads menu for easy access to DNA sequences of several other industrial strains and environmental isolates. The new Sequence, Chromosome, and Contig pages make use of many of the features you enjoy on other new or redesigned pages at SGD, including graphical display of data, sortable tables, and responsive visualizations. The Sequence pages also provide seamless access to other tools at SGD such as BLAST and Web Primer. Please explore these new pages, accessible via the Sequence tab on your favorite Locus Summary page, and send us your feedback.
New fungal homolog data at SGD
Have you ever wondered about the role played by the homolog of a particular yeast gene in other fungal species? SGD’s advanced search tool, YeastMine, can now be used to find homologs of your favorite Saccharomyces cerevisiae genes in the pathogenic yeast, Candida glabrata. There are now 25 species of pathogenic and non-pathogenic fungi in YeastMine, including S. cerevisiae.
The fungal homologs of a given S. cerevisiae gene can be found using the template called “Gene –> Fungal Homologs.” Fungal homology data comes from various sources including FungiDB, the Candida Gene Order Browser (CGOB), the Yeast Gene Order Browser (YGOB), the Candida Genome Database (CGD), the Aspergillus Genome Database (AspGD) and PomBase, and the results link directly to the corresponding homolog gene pages in the relevant databases.
A results table is generated after each query and the identifiers and standard names for the fungal homologs are listed in the table. As with other YeastMine templates, results can be saved as lists for further analysis. You can also create a list of yeast gene names and/or identifiers using the updated Create Lists feature that allows you to specify the organism representing the genes in your list. The query for homologs can then be made against the custom gene list.
All of the new templates that query fungal homolog data can be found on the YeastMine Home page under the “Homology” tab. This template complements the template “Gene → Non-Fungal and S. cerevisiae Homologs” that retrieves homologs of S. cerevisiae genes in humans, rats, mice, worms, flies, mosquitos, and zebrafish.
We invite you to watch SGD’s YeastMine Fungal Homologs video tutorial for tips on accessing Fungal Homolog data at SGD. You can view all Video Tutorials for YeastMine here.
SGD at the Yeast Genetics Meeting
In late July, SGD staff members traveled to the University of Washington in Seattle to attend the premier meeting of the yeast research community: the Yeast Genetics Meeting, organized by the Genetics Society of America. This conference brought together almost 600 researchers and educators with an interest in Saccharomyces cerevisiae and other yeasts.
SGD’s table at the poster sessions was a popular gathering spot for discussion about SGD and hands-on demonstrations. We also presented a well-attended and lively workshop session on several different SGD tools and resources. And we were very gratified to receive a round of applause from conference attendees at the final banquet, when keynote speaker Anita Hopper publicly thanked SGD’s P.I. Mike Cherry and staff for our service to the yeast community.
We chronicled the meeting in tweets, and you can view them by searching Twitter with the hashtag #YEAST14. In addition, check out this entertaining, photo-filled recap of the meeting from our friends at GSA.
 Selina's presentation on YeastMine |
Upcoming Conferences and Meetings
- Rennes, France
- October 24, 2014
- Pacific Grove, CA
- March 17-22, 2015
- Abstract deadline: December 10, 2014
- Beijing, China
- April 23-26, 2015
- Abstract deadline: February 1, 2015
- Levico Terme, Italy
- September 7-12, 2015
- Orlando, FL
- July 13-17, 2016
*asterisks indicate attendance by SGD
Note: If you no longer wish to receive this newsletter, please contact the SGD Help Desk at sgd-helpdesk@lists.stanford.edu.