SGD Newsletter, March 2016
About this newsletter:
This is the March 2016 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community.
You can also subscribe to SGD's RSS feed to receive updates on SGD news:
http://www.yeastgenome.org/feed
Contents
Introducing the SGD Webinar Series: April 6
SGD: A Catalyst for Biological Research
Function is just one of many important angles to consider a gene and its product. Other concepts like regulation, genetic/physical interactions, phenotypes, are critical to understanding how a gene fits into the overall biology of the cell. By quickly connecting users to high-quality annotations, different types of data, and useful tools to explore the yeast genome, SGD acts as a catalyst for biological research and has become an indispensable resource for many scientists.
We are excited to announce our first webinar of the SGD Webinar Series on April 6th, 2016, 9:30 AM PDT. In this short webinar, we will provide an overview of the different ways you can find information about your favorite genes in SGD.
Space for this webinar is limited. To reserve your spot, please register for this event here: http://bit.ly/229od64
SGD at The Allied Genetics Conference (TAGC) 2016
SGD will be attending The Allied Genetics Conference (TAGC) in Orlando, Florida, July 13–17, 2016! For the first time ever, the meetings of the yeast, C. elegans, ciliate, Drosophila, mouse, and zebrafish model organism communities will be united under one roof, along with a new meeting on population, evolutionary, and quantitative genetics.
Submit your abstracts now! Abstract submission will be open until March 23, 2016. If you want GREAT science and access to the leaders of the field, then TAGC is the place for you. SGD will be there, will you?
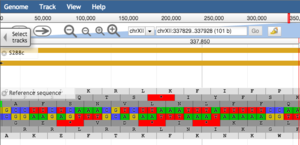
Coming Soon to SGD: Updated Genome Browser
SGD will soon be releasing an updated genome browser to enable fast and responsive exploration of the S. cerevisiae genome. Based on JavaScript and HTML5, JBrowse offers a smooth user experience with continuous page transitions, quick zooming, and a clean interface with ample room to visualize data tracks. Take a sneak peek at www.yeastgenome.org/browse and send us comments or suggestions.
Keep an eye out for the updated genome browser to go live on SGD later this month.
Research Spotlight (Blog Posts)
In case you missed them, here are some of the Research Spotlights posted on our blog lately:
- Unlocking Chromatin The yeast gene HO is notoriously difficult to activate. It requires three regulatory "locks" to be opened in order to increase its expression. Yarrington et al. found that the second "lock" is a set of nucleosomes, and that their absence at URS2-L resulted in fewer steps required for HO activation.
- Unfrying An Egg Almost like un-frying an egg, yeast cells were shown by Wallace et al. to have the ability to restore aggregated proteins back to their normal shape after a heat shock.
- Of Medieval Market Townes and Wasp Guts A secret hookup spot for yeast to mate has been discovered by Stefanini et al. in a place that really sets the mood: the wasp gut.
- Not Quite The Same Yeast that bake their own "bread", or make their own nutrients, work very differently from yeast that can't and are instead provided an abundant supply of nutrition, as Alam et al. found.
- The Benefits of Sex If yeast can reproduce asexually, why bother with sex? McDonald et al. found that over time, sexual yeast populations "weed out" unimportant mutations while keeping the ones that actually make a difference.
- Clearing Customs in the Nucleus Much like going through airport customs, mRNAs trying to exit the nucleus are checked before entering the cytoplasm. Smith et al. and Saroufim et al. found ways to study this fascinating process of mRNA export in real-time.
- Budding Yeast Diversifies its Phosphatase Portfolio Have you ever heard the saying, "Don't put all your eggs in one basket"? It turns out the budding yeast S. cerevisiae may be wiser than the fission yeast S. pombe, as Kennedy and coworkers discovered that our favorite budding yeast has three different phosphatases that can activate Cdk1 and promote mitosis instead of one.
Recent Publications from SGD
- Cherry JM (2015) The Saccharomyces Genome Database: A Tool for Discovery. Cold Spring Harb Protoc. 2015. Dec 2;2015(12):pdb.top083840. doi: 10.1101/pdb.top083840. PMID: 26631132
- Cherry JM (2015) The Saccharomyces Genome Database: Exploring Genome Features and Their Annotations. Cold Spring Harb Protoc. 2015 Dec 2;2015(12):pdb.prot088922. doi: 10.1101/pdb.prot088922. PMID:26631126
- Cherry JM (2015) The Saccharomyces Genome Database: Gene Product Annotation of Function, Process, and Component. Cold Spring Harb Protoc. 2015 Dec 2;2015(12):pdb.prot088914. doi: 10.1101/pdb.prot088914. PMID: 26631125
- Cherry JM (2015) The Saccharomyces Genome Database: Advanced Searching Methods and Data Mining. Cold Spring Harb Protoc. 2015 Dec 2;2015(12):pdb.prot088906. doi: 10.1101/pdb.prot088906. PMID: 26631124
- Cherry JM (2015) The Saccharomyces Genome Database: Exploring Biochemical Pathways and Mutant Phenotypes. Cold Spring Harb Protoc. 2015 Dec 2;2015(12):pdb.prot088898. doi: 10.1101/pdb.prot088898. PMID: 26631123
Upcoming Meetings
- University of Alabama, Tuscaloosa, AL, USA
- March 25-27, 2016
- Registration and abstract submission closed
- Campus Biotech, Geneva, Switzerland
- April 10-14, 2016
- Registration deadline: April 1, 2016
- Abstract submission closed
- Brussels, Belgium
- April 13-15, 2016
- Registration and abstract submission deadline: February 16, 2016
- Late registration and abstract submission still possible
- Sant Feliu de Guixols, Spain
- June 11-16, 2016
- Registration and abstract submission deadline: March 14, 2016
- University of Lisbon, Lisbon, Portugal
- July 11-14, 2016
- Abstract and early registration deadline: March 15, 2016
- Registration deadline: July 1, 2016
- Orlando, FL, USA
- July 13-17, 2016
- Abstract and early registration deadline: March 23, 2016
- Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- July 26 - August 15, 2016
- Application Deadline: April 15, 2016
- Snowmass, CO, USA
- July 31-August 5, 2016
- Early Bird deadline: June 16, 2016
- Last day to register: July 1, 2016
- Awaji Yumebutai International Conference Center, Hyogo, Japan
- September 11-15, 2016
- Abstract submission and early registration deadline: June 10, 2016