SGD Newsletter, December 2016
About this newsletter:
This is the December 2016 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community.
Contents
- 1 New Phenotype Data
- 2 New JBrowse Tracks
- 3 New Protein Half-life Data
- 4 SGD Webinar Series
- 5 SGD Help Videos
- 6 The ALLIANCE of Genome Resources
- 7 Another Nobel Prize for Yeast!
- 8 Research Spotlight (Blog posts)
- 9 SGD swag: out and about
- 10 Recent Publications from SGD Staff
- 11 Happy Holidays from SGD!
- 12 Upcoming Meetings
New Phenotype Data
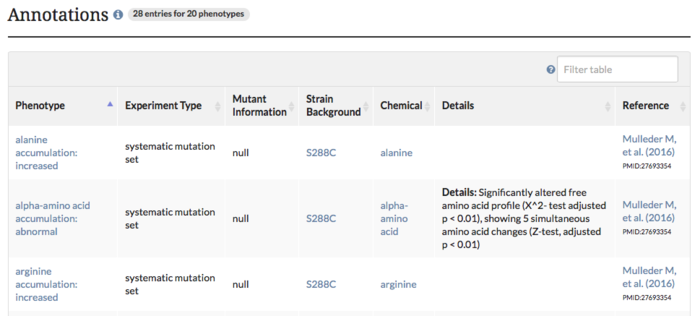
4217 new phenotypes for 1520 genes have been added to SGD from Mülleder et al. They determined the free amino acid concentration profiles of null mutants from the systematic knockout set in S288C to see which genes have similar profiles. In addition to adding these phenotypes, we've also added links on the Phenotype pages to the Metabolic Gene Card for each ORF that has a profile. For example, ACO2/YJL200C has increased arginine, tyrosine and alanine accumulation and decreased proline and lysine accumulation (see above image). These are all found within the table on the Phenotype page. The MetaboGeneCard link in the "Resources" section will show other genes with similar profiles.
Thank you to Michael Mülleder for his help in preparing this large dataset.
New JBrowse Tracks
New tracks have been added to SGD's instance of JBrowse. Ostrow et al's genome-wide study of Fkh1p and Fkh2p chromatin binding sites is now available in JBrowse. Sun et al's genome-wide study of axis protein binding to chromosomes is now also available for visualization on JBrowse. All track files are available for download in the Published Datasets directory of our downloads site.
We keep adding new tracks to JBrowse for visualization. Please let us know of any publicly available datasets that you think should be added!
New Protein Half-life Data
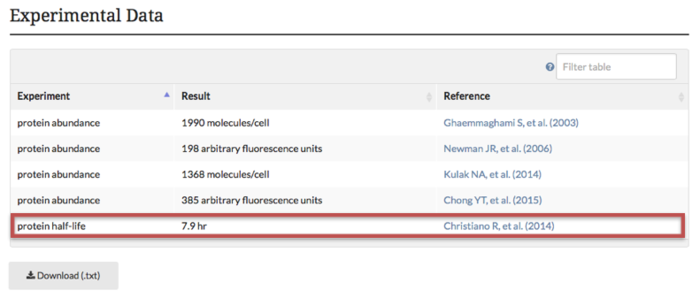
Protein half-life measurements are now available in the "Experimental Data" section of our protein tab pages (see example above from STH1 protein tab). The protein half-life data is based on data from a study by Christiano et al. (2014). Additionally, this half-life data can also be retrieved using YeastMine templates Gene-->Protein Half-life and Retrieve-->Proteins with half-life in a given range . Both of the YeastMine half-life templates are accessible from the “templates” section under the “protein” category.
SGD Webinar Series
SGD is hosting webinars to help you, our users, learn more about useful SGD tools, see expanded research examples, and ask SGD biocuration scientists questions live. The latest webinar in our webinar series was on December 14, 2016 and discussed how to use SGD to find human/yeast homologs and their disease implications. If you are interested in this topic, but were unable to attend the webinar, watch the recorded version here!
To read about and register for SGD's latest webinars, or to browse the videos and materials of past webinars, visit our SGD Webinars page.
Check out our latest recorded webinars:
- SGD Webinars: Homology and Disease Data at SGD
- SGD Webinars: Exploring Expression Datasets & Coexpressed Genes with SPELL
SGD Help Videos
SGD is actively expanding our library of short video tutorials to help you use various SGD tools and pages. Check out our latest help videos:
- SGD Help: Finding Human Homology & Disease Information
- SGD Help: Exploring Expression Datasets with SPELL
Videos are accessible via SGD's YouTube channel. If you have a great idea for a video, or would like to see a particular topic covered, feel free to contact us!
The ALLIANCE of Genome Resources
Work on the collaboration with WormBase, FlyBase, ZFIN, MGI, RGD, and the Gene Ontology Consortium to establish the Alliance of Genome Resources is well under way! Scientists from all groups are coming together to develop the best integrated tools for our users and to provide users with easy access to all the data from the Model Organism Databases.
For more updates and announcements on the Alliance, check out the Alliance Facebook page, or check out our twitter page @alliancegenome.
Another Nobel Prize for Yeast!
In October Dr. Yoshinori Ohsumi won the 2016 Nobel Prize in Physiology or Medicine for his work on autophagy in yeast. Dr. Ohsumi used Sacchamomyces cerevisiae as a model organism to make discoveries about the autophagy mechanism which has important human disease applications. Read more about it here! Behold the #APOYG!
Research Spotlight (Blog posts)
In case you missed them, here are some of the most popular Research Spotlights posted on our blog lately:
- A Biological Tour de Force Reveals the Complexity of a Yeast Cell Costanzo and colleagues constructed more than 23 million double mutants in order to generate a global genetic interaction network which will provide information to researchers for years to come. Explore these interactions at http://thecellmap.org
- Winter is Coming (for Cancer) Reid and coworkers used a yeast synthetic dosage lethality assay to find new potential cancer targets.
- Personalized Essential Genes Rowley and coworkers demonstrated that while XRN1 could be swapped across four different Saccharomyces species and maintain basic function, personalized XRN1 genes were much better at protecting their own species against species-specific viruses than the XRN1 genes from other species.
- Too Much of a Good Thing Ang and coworkers investigated which genes might have an increased mutation rate when they are overexpressed - yet another example of how yeast can be a powerful tool in human disease research!
- Yeast Tackles Climate Change Sato and coworkers conducted evolution experiments in order to investigate the genes involved in yeast that was able to grow on only xylose, which has implications in biofuel production.
SGD swag: out and about
Do you have pictures to share of SGD swag out and about? Send them to SGD curators for possible inclusion in a future newsletter!
Recent Publications from SGD Staff
1. Engel, S.R., and MacPherson, K.A. 2016. Using model organism databases (MODs). Curr. Protoc. Essential Lab. Tech. 13:11.4.1-11.4.22. doi: 10.1002/cpet.4
Happy Holidays from SGD!
We want to take this opportunity to wish you and your family, friends and lab mates the best during the upcoming holidays. Stanford University will be closed for two weeks starting on December 21st, reopening on January 4th, 2017. Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will attempt to keep connected via email should you have any questions.
Upcoming Meetings
24th Annual Southeastern Regional Yeast Meeting (SERYM 2017)
Hotel Capstone Conference Center, University of Alabama, Tuscalossa, AL
March 3-5, 2017
29th Fungal Genetics Conference
Asilomar Conference Center, Pacific Grove, CA
March 14-19, 2017
Abstract submission and registration deadline: December 8, 2016
9th INTERNATIONAL FISSION YEAST MEETING
The Banff Centre, Banff, Alberta, Canada
May 14-19, 2017
Abstract submission deadline: February 17, 2017
Registration deadlines: December 1, 2016 (Early bird), February 15, 2017 (regular) and April 1, 2017 (late)
12th International Meeting on Yeast Apoptosis
Camera di Commercio di Bari, Bari, Italy
May 14-18, 2017
13th Yeast Lipid Conference
AgroParisTech Claude Bernard campus, Paris, France
May 17-19, 2017
ISSY33: Exploring and Engineering Yeasts for Industrial Application
University College Cork, Ireland
June 26-29, 2017
28th International Conference on Yeast Genetics and Molecular Biology (ICYGMB)
Prague Congress Centre, Prague, Czech Republic
August 27 – September 1, 2017