YJL133C-A
Share your knowledge...Edit this entry! <protect>
| Systematic name | YJL133C-A |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr X:159847..159623 |
| Primary SGDID | S000028805 |
Description of YJL133C-A: Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies[1]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
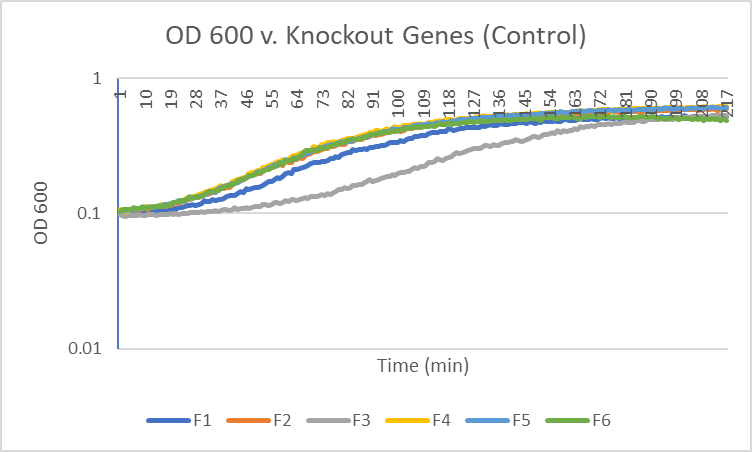
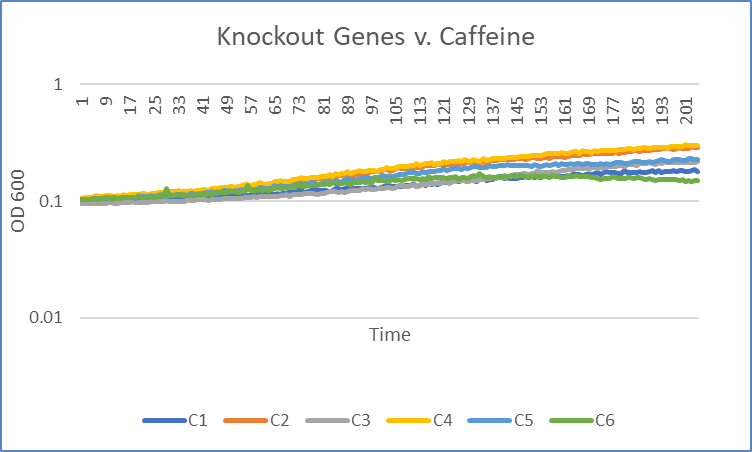
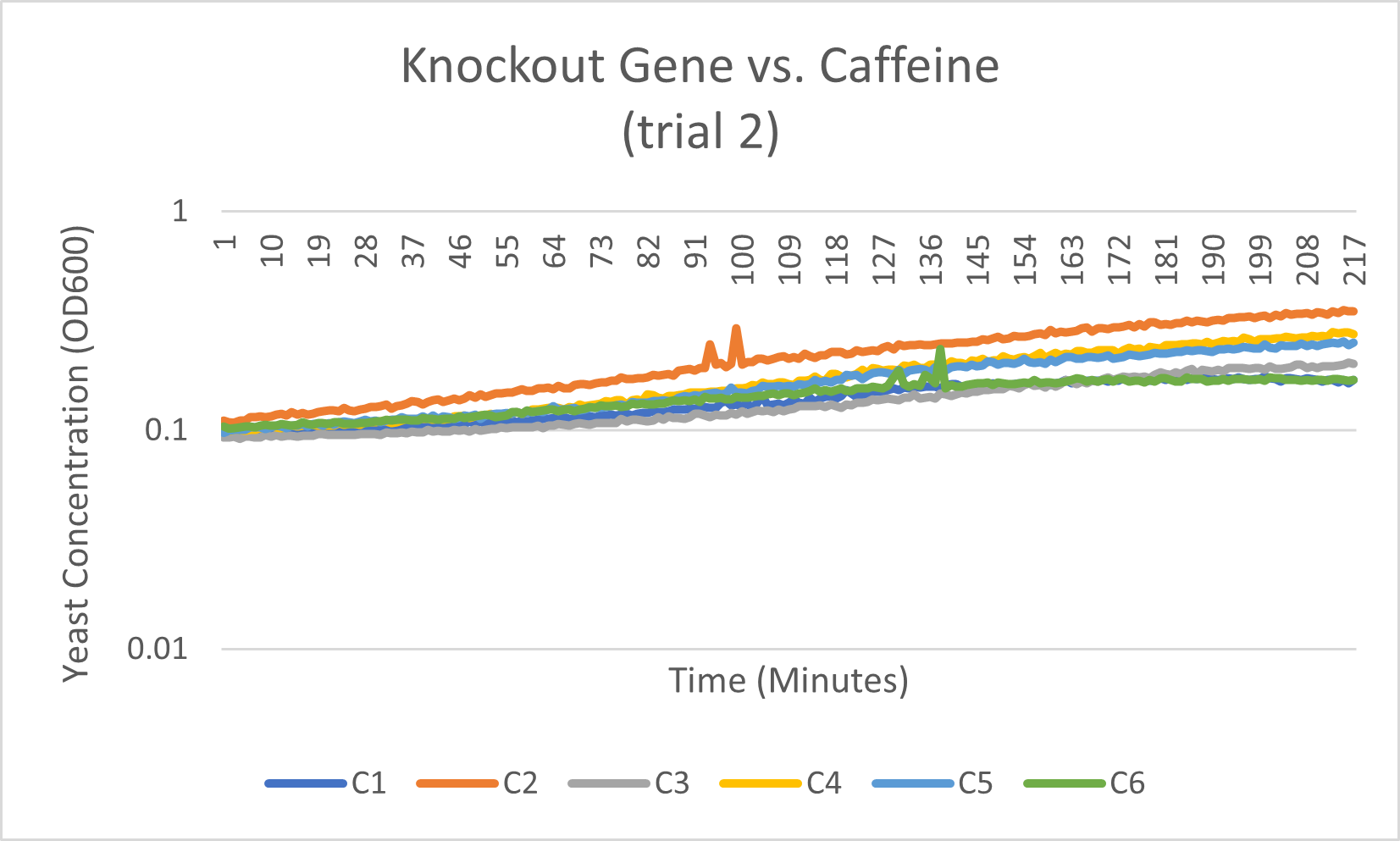
Caffeine
Interpretation
The average doubling time for the YJL133C-A (C2) gene in standard conditions was 89.5 minutes. Under caffeine stress, the average doubling time was 134 minutes. That is a 49.7% increase in doubling time which means that cell growth was negatively affected (slowed down) by this type of stress.
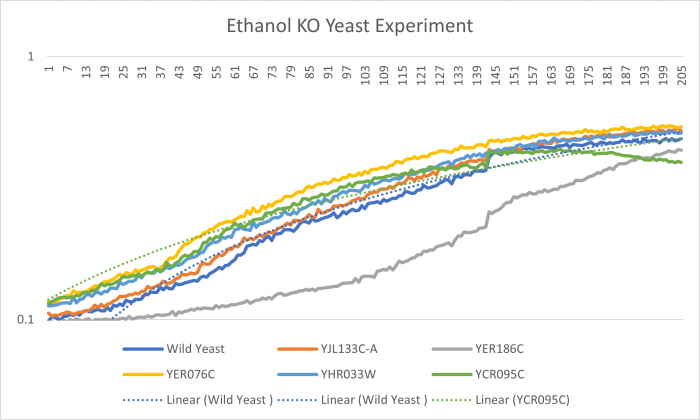
Ethanol
YJL133C-A KO gene
UV Light
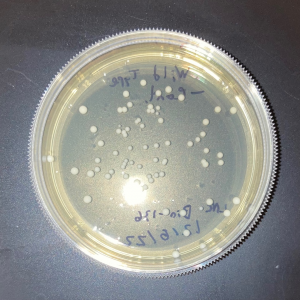
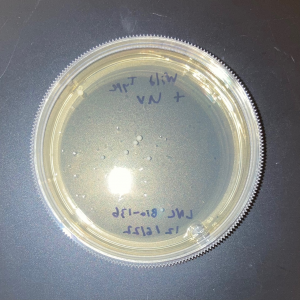
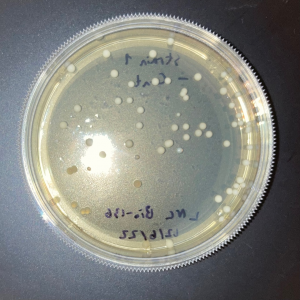
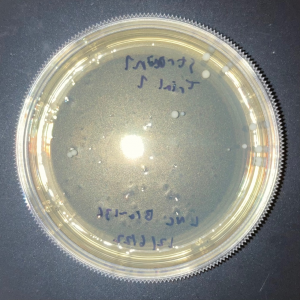
The plate with the modified yeast that was not exposed to UV had approximately 5% fewer colonies than the wild-type plate that was not exposed to UV. This shows that the knock-out strain of yeast is expected to be less viable compared to wild-type yeast in the absence of stressors.
The two trial plates were pretty similar in colony count. The average of the two was 10 colonies.
The plate with wild-type yeast that was exposed to UV had 11 colonies.
There was no significant difference in colony count between the modified yeast and wild-type yeast that was exposed to UV.
As there was no significant difference between the wild-type yeast exposed to UV and the trial strains, we can conclude that the gene is not used in the defense against UV damage nor the repair of UV damage in yeast.
<protect>
References
See Help:References on how to add references
See Help:Categories on how to add the wiki page for this gene to a Category </protect>
UW Stout/FA 2022 Glycerol Media
Introduction
Following the UW-Stout/Glycerol FA22 protocol, 5 knockout strains of yeast cell were tortured within a glycerol media solution.
Results
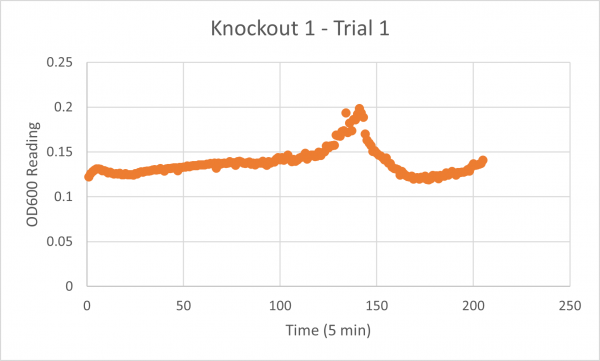
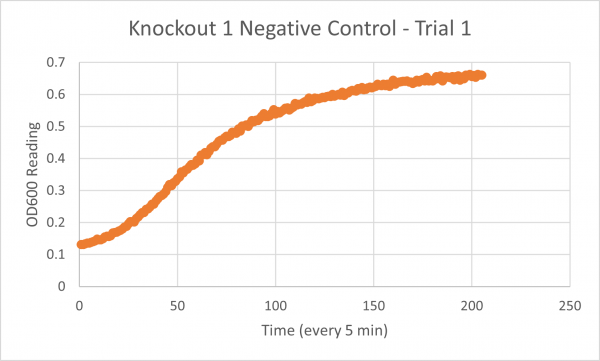
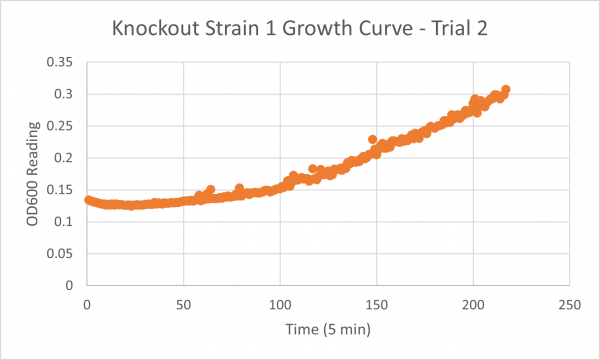
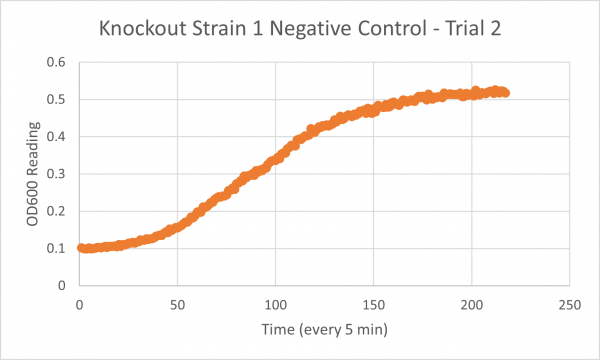
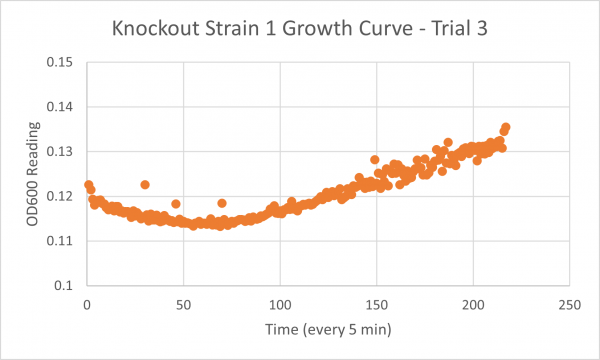
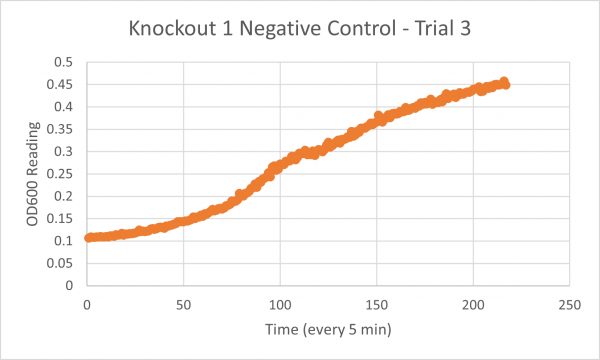 The following doubling times were calculated:
The following doubling times were calculated:
- Negative Control Trial 1: 163 minutes
- Glycerol Trial 2: 565 minutes
- Negative Control Trial 2:
- Glycerol Trial 3:
- Negative Control Trial 3: