Difference between revisions of "YER076C"
SGDwikiBot (talk | contribs) (Automated import of articles) |
|||
| (31 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
{|{{Prettytable}} align = 'right' width = '200px' | {|{{Prettytable}} align = 'right' width = '200px' | ||
|- | |- | ||
| − | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http:// | + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http://www.yeastgenome.org/cgi-bin/locus.pl?dbid=S000000878 YER076C] |
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | ||
| − | |nowrap| Chr V: | + | |nowrap| Chr V:313498..312590 |
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Primary SGDID''' || S000000878 | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Primary SGDID''' || S000000878 | ||
|} | |} | ||
<br> | <br> | ||
| − | '''Description of YER076C:''' Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; analysis of HA-tagged protein suggests a membrane localization<ref name='S000117178'>Reinders J, et al. (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5(7):1543-54 {{SGDpaper|S000117178}} PMID 16823961</ref><ref name=' | + | '''Description of YER076C:''' Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; analysis of HA-tagged protein suggests a membrane localization<ref name='S000117178'>Reinders J, et al. (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5(7):1543-54 {{SGDpaper|S000117178}} PMID 16823961</ref><ref name='S000075100'>Sickmann A, et al. (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A 100(23):13207-12 {{SGDpaper|S000075100}} PMID 14576278</ref><ref name='S000075303'>Terashima H, et al. (2002) Sequence-based approach for identification of cell wall proteins in Saccharomyces cerevisiae. Curr Genet 40(5):311-6 |
| − | {{SGDpaper| | + | {{SGDpaper|S000075303}} PMID 11935221</ref> |
<br> | <br> | ||
<br> | <br> | ||
| Line 30: | Line 30: | ||
{{CommentaryHelp}} | {{CommentaryHelp}} | ||
| + | ===[[UW-Stout/Ethanol_FA22|Ethanol]]=== | ||
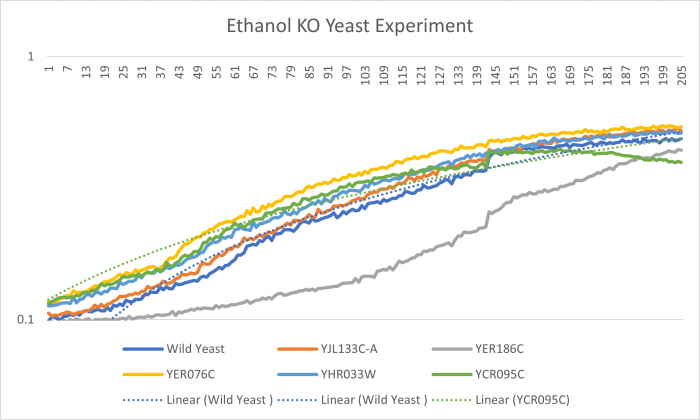
| + | [[Image:Best Ethanol KO graph.png|Graph|center|700px|YJL133C-A]] | ||
| + | YER076C had the most cell growth. The doubling time was 71.60. Ethanol didn't affect the growth to much. | ||
| + | ===[[UW-Stout/Caffeine_FA22|Caffeine]]=== | ||
| + | |||
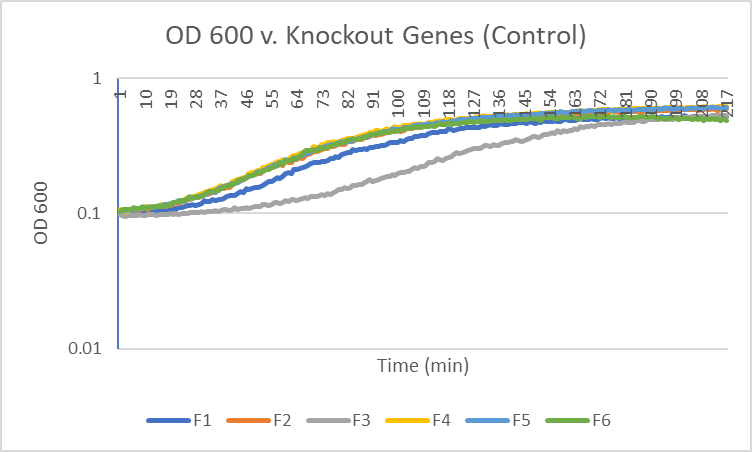
| + | [[Image:F_graph.png|100pxs|Control]] | ||
| + | |||
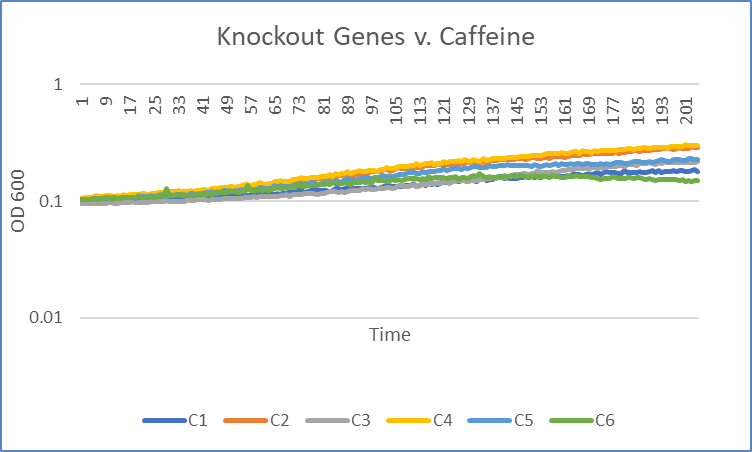
| + | [[Image:Knockout_v._caffeine_trial1_Viv.png|100pxs|Trial 1]] | ||
| + | |||
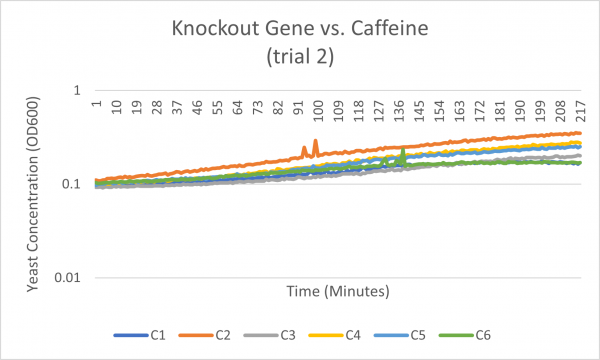
| + | [[Image:Knockout_gene_v._Caffeine_trial_2.png|600px|Trial 2]] | ||
| + | |||
| + | ====Interpretation==== | ||
| + | |||
| + | The average doubling time for the YER076C (C4) gene in standard conditions was 87.5 minutes. Under caffeine stress, the average doubling time was 139.5 minutes. That is a 59.4% increase in doubling time which means that cell growth was negatively affected (slowed down) by this type of stress. | ||
| + | |||
| + | ===Hydrogen Peroxide=== | ||
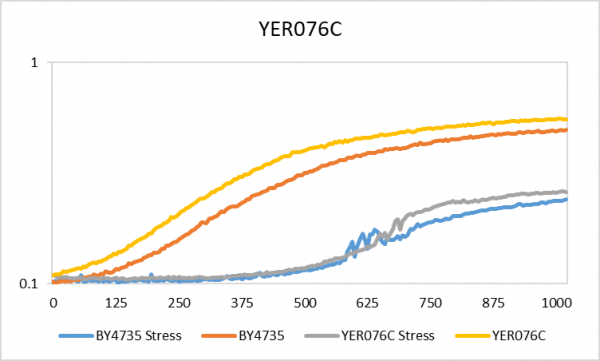
| + | [[Image:YER076C.png|center|thumb|600px|The x-axis is time in minutes and the y-axis is OD600]] | ||
| + | This is the link to the protocol page: [[UW-Stout/Hydrogen Peroxide FA22]]. | ||
| + | |||
| + | The YER076C gene under the hydrogen peroxide media had a doubling time of 358 minutes whereas with no stress a doubling time of 210 minutes. The BY4375 under stress had a 598 minute doubling time and with no stress there was a 229 minute doubling time. Based on the chart above and the doubling times, no large effect when the YER076C gene is knocked out. | ||
<!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | <!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | ||
| Line 43: | Line 63: | ||
<protect> | <protect> | ||
| + | |||
==References== | ==References== | ||
<!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | <!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | ||
{{RefHelp}} | {{RefHelp}} | ||
</protect> | </protect> | ||
| + | |||
| + | ==UW Stout/FA 2022 Glycerol Media== | ||
| + | ===Introduction=== | ||
| + | Following the [[UW-Stout/Glycerol FA22]] protocol, 5 knockout strains of yeast cell were tortured within a glycerol media solution. | ||
| + | ===Results=== | ||
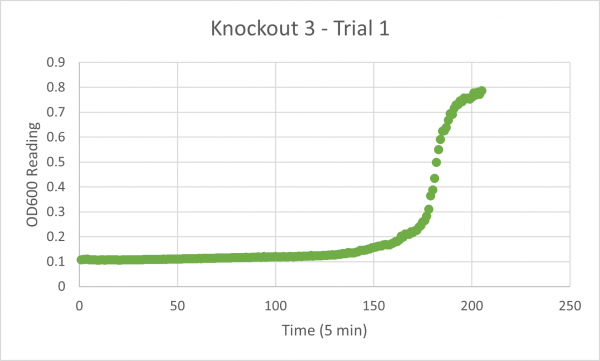
| + | [[Image:Knockout_3_Trial_1.png|600px]] | ||
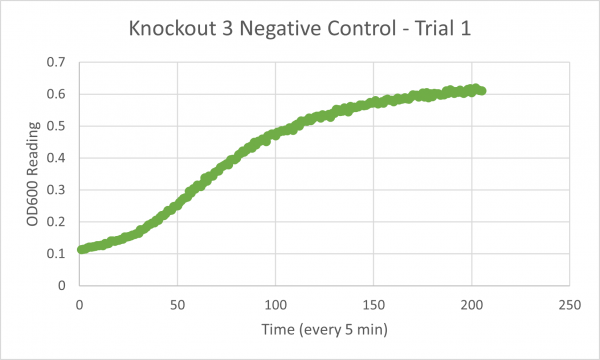
| + | [[Image:Negative Control KO3 Trial 1.png|600px]] | ||
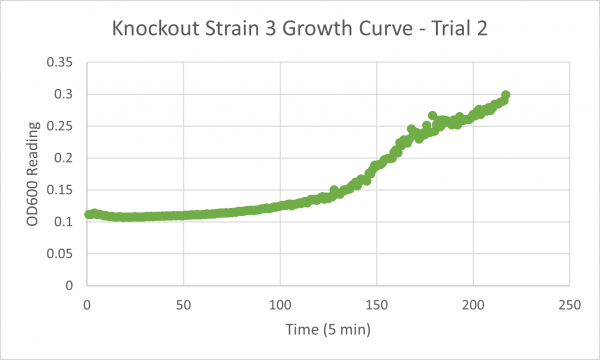
| + | [[Image:Knockout_3_Trial_2.png|600px]] | ||
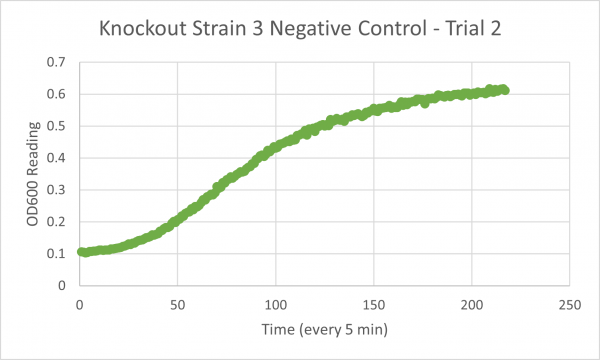
| + | [[Image:Negative_Control_KO3_Trial_2.png|600px]] | ||
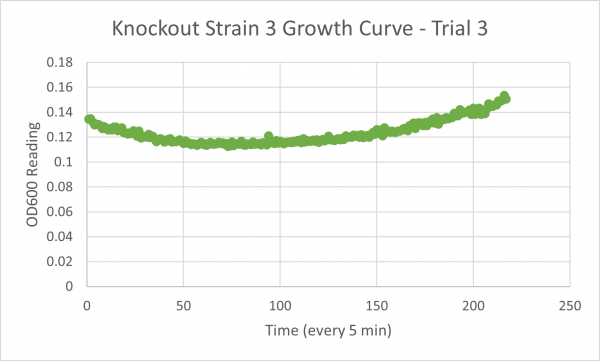
| + | [[Image:Growth_Curve_KO3_Trial_3.png|600px]] | ||
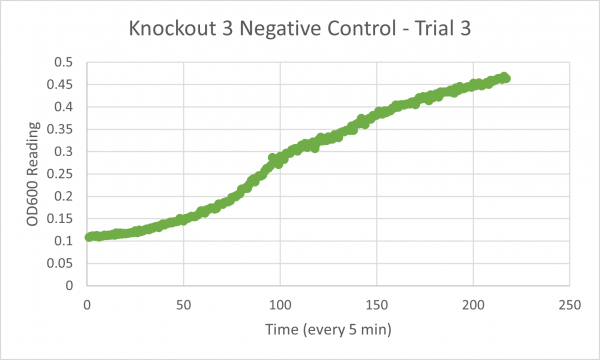
| + | [[Image:Negative_Control_KO3_Trial_3.png|600px]] | ||
| + | |||
| + | The following doubling times were calculated: | ||
| + | * Glycerol Trial 1: NA | ||
| + | * Negative Control Trial 1: 188 min | ||
| + | * Glycerol Trial 2: 144 min | ||
| + | * Negative Control Trial 2: 192 min | ||
| + | * Glycerol Trial 3:2531 min | ||
| + | * Negative Control Trial 3: 212 min | ||
| + | |||
| + | ===Analysis/Conclusion=== | ||
| + | Our results for our glycerol media is not consistent, the first trial was messed up, and the second and third trial have a large difference. Trials 2 & 3 bring up some issues because trial 2's time is lower than the negative control which would mean that this knockout gene strain of yeast would work better in the glycerol media than the glucose media, but our time for trial 3 is much larger than the negative control, and because our first trial was inconclusive we would need to run another trial to figure out whether or not this strain would truly live better in the glycerol media or not. | ||
| + | |||
| + | =UW Stout/FA 2022 Bud Scars= | ||
| + | Cells that had the gene YER076C had an average of 1.46 scars per cell. Out of 100 cells counted, 146 bud scars were counted. The wild yeast BY4735 had an average of 2.46 bud scars per cell (100 cells counted, 246 bud scars). This indicates that the knocked-out gene significantly slowed down the reproduction rate of these cells. | ||
| + | |||
| + | [[File:Yeast_3(2)_2_53.png]] | ||
| + | ''picture does not include all cells that were counted and added for a visual of bud scars'' | ||
Latest revision as of 18:41, 20 December 2022
Share your knowledge...Edit this entry! <protect>
| Systematic name | YER076C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr V:313498..312590 |
| Primary SGDID | S000000878 |
Description of YER076C: Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; analysis of HA-tagged protein suggests a membrane localization[1][2][3]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
Ethanol
YER076C had the most cell growth. The doubling time was 71.60. Ethanol didn't affect the growth to much.
Caffeine
Interpretation
The average doubling time for the YER076C (C4) gene in standard conditions was 87.5 minutes. Under caffeine stress, the average doubling time was 139.5 minutes. That is a 59.4% increase in doubling time which means that cell growth was negatively affected (slowed down) by this type of stress.
Hydrogen Peroxide
This is the link to the protocol page: UW-Stout/Hydrogen Peroxide FA22.
The YER076C gene under the hydrogen peroxide media had a doubling time of 358 minutes whereas with no stress a doubling time of 210 minutes. The BY4375 under stress had a 598 minute doubling time and with no stress there was a 229 minute doubling time. Based on the chart above and the doubling times, no large effect when the YER076C gene is knocked out.
<protect>
References
See Help:References on how to add references
- ↑ Reinders J, et al. (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5(7):1543-54 SGD PMID 16823961
- ↑ Sickmann A, et al. (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A 100(23):13207-12 SGD PMID 14576278
- ↑ Terashima H, et al. (2002) Sequence-based approach for identification of cell wall proteins in Saccharomyces cerevisiae. Curr Genet 40(5):311-6 SGD PMID 11935221
See Help:Categories on how to add the wiki page for this gene to a Category </protect>
UW Stout/FA 2022 Glycerol Media
Introduction
Following the UW-Stout/Glycerol FA22 protocol, 5 knockout strains of yeast cell were tortured within a glycerol media solution.
Results
The following doubling times were calculated:
- Glycerol Trial 1: NA
- Negative Control Trial 1: 188 min
- Glycerol Trial 2: 144 min
- Negative Control Trial 2: 192 min
- Glycerol Trial 3:2531 min
- Negative Control Trial 3: 212 min
Analysis/Conclusion
Our results for our glycerol media is not consistent, the first trial was messed up, and the second and third trial have a large difference. Trials 2 & 3 bring up some issues because trial 2's time is lower than the negative control which would mean that this knockout gene strain of yeast would work better in the glycerol media than the glucose media, but our time for trial 3 is much larger than the negative control, and because our first trial was inconclusive we would need to run another trial to figure out whether or not this strain would truly live better in the glycerol media or not.
UW Stout/FA 2022 Bud Scars
Cells that had the gene YER076C had an average of 1.46 scars per cell. Out of 100 cells counted, 146 bud scars were counted. The wild yeast BY4735 had an average of 2.46 bud scars per cell (100 cells counted, 246 bud scars). This indicates that the knocked-out gene significantly slowed down the reproduction rate of these cells.
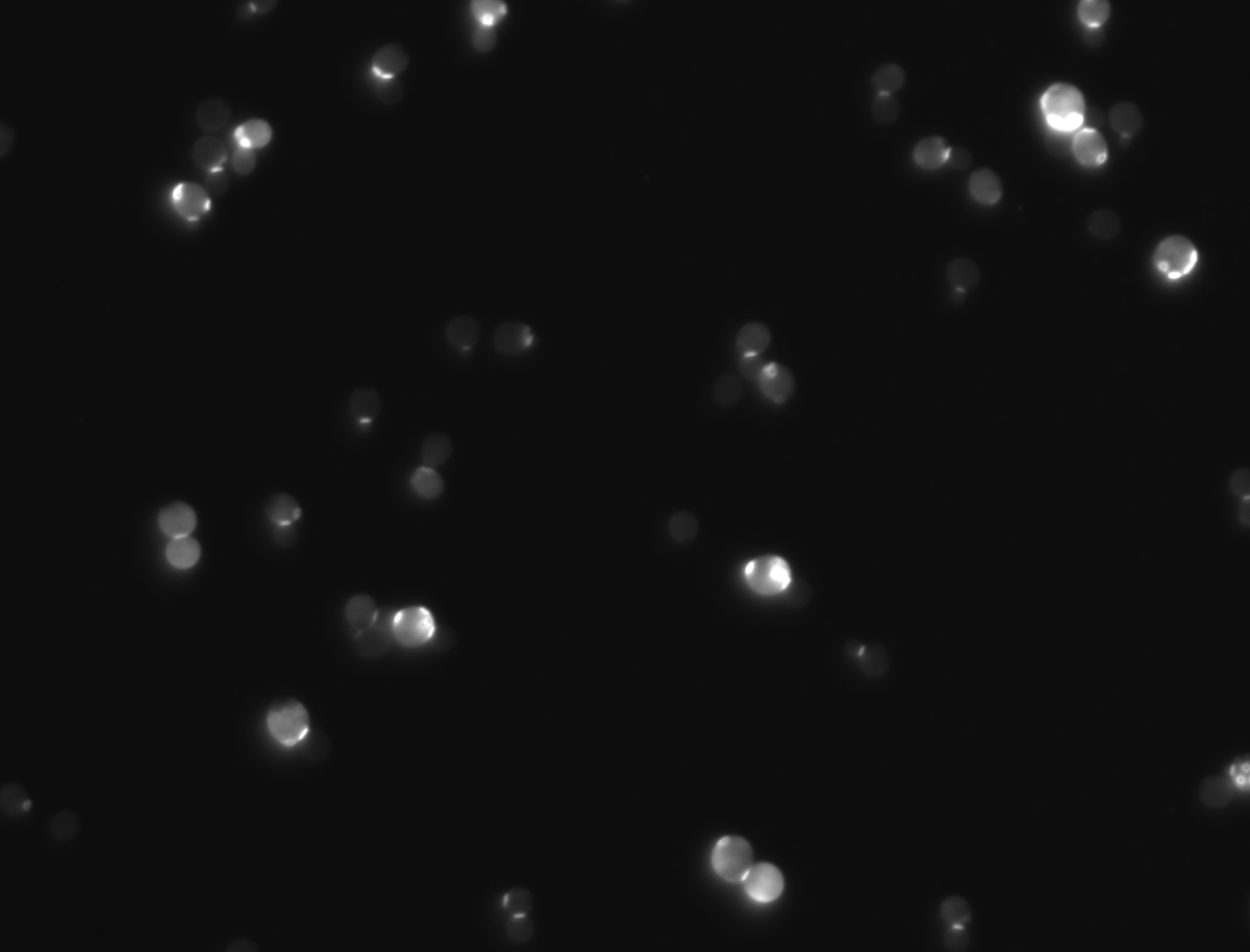 picture does not include all cells that were counted and added for a visual of bud scars
picture does not include all cells that were counted and added for a visual of bud scars