Difference between revisions of "UW-Stout/Heat Shock FA21"
(→Important Remaining Items (Remove Before Publish)) |
(→Important Remaining Items (Remove Before Publish)) |
||
| Line 177: | Line 177: | ||
| − | You repeated the same image on each gene page -- that's fine, but I can't tell which plate has which strain or condition on it! | + | 1). You repeated the same image on each gene page -- that's fine, but I can't tell which plate has which strain or condition on it! Which means that interpreting your data is basically impossible. |
| − | + | 2). Additionally, you present an image that's the result of your calibration experiment -- but you never tell me what I'm looking at or what it means. | |
| − | Finally, the details of the calibration protocol -- for how much time you shocked your cells -- seem wrong. Didn't you load them all in at the same time, then take them out at particular timepoints? | + | 3). Finally, the details of the calibration protocol -- for how much time you shocked your cells -- seem wrong. Didn't you load them all in at the same time, then take them out at particular timepoints? |
That's not what the protocol says. Also, please include the serial dilution that you did in your protocol! | That's not what the protocol says. Also, please include the serial dilution that you did in your protocol! | ||
Revision as of 12:04, 20 December 2021
Contents
Materials
96 channel well
Yeast cells
PCR test tubes
Media and Plates (all in 475ml H2O)
500 ul of YPD
10g Peptide
5g Yeast Extract
10g Agarose
Equipment
Thermocycler
Incubator at 30°C
P20 and P200 micro pipettes
Multichannel pipette
Protocol Page
Calibration Experiment
Heat Shock Procedure
- Set thermocycler to 48°C sitting temperature.
- Vortex the yeast culture briefly to resuspend the yeast cells.
- Label 8 pcr tubes A through H.
- Micropipette 90 ul of water into the pcr tubes.
- Micropipette 10 ul of a strain
- Label each tube “A, B, C, D, E, F, G, H”.
- Insert all 8 of the tubes into the thermocycler
- Remove a tube every 8 minutes and 30 seconds for an 1 hour from the thermocycler at 48°C.
Dilution Procedure
- In a 96 channel well put in your six yeast strains then put them into the left most column descending from A to F.
- Then in the rest of the rows add 90 ul of H2O into A-H.
- After adding the water take 10 ul of the strains add them to the next column and mix.
Plating Procedure
- Label each petri dish accordingly, first plate "A,B", the Second plate "C,D", the Third plate "E,F" and the fourth plate "G,H".
- Micropipette 5 ul of the strains onto the coordinated petri dish using the .
- Place all the petri dishes in the incubator (30°C).
- Let sit for 48 hours.
- Take out and analyze the growth.
- Repeat for 3 trials.
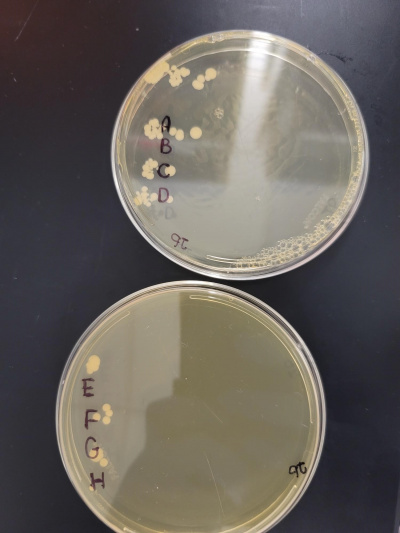
Raw Data
Interpretation
A: It was the control with 0 minutes heat shocked.
B: It was in for 8 minutes and 30 seconds.
C: Was in for 17 minutes.
D: It was in for 25 minutes and 30 seconds.
E: Was in for 33 minutes.
F: It was in for 41 minutes and 30 seconds.
G: Was in for 50 minutes.
H: It was in for 58 minutes and 30 seconds.
From what we gathered we found out that over time the cells died off and at around 42 minutes it stressed them the most but did not kill them.
Final Protocol
Media Creation Procedure
- Measure out 10g of Peptide and put into 500ml jar.
- Measure out 5g of Yeast Extract and put into 500ml jar.
- Measure out 10g of Agarose and put into 500ml jar.
- Then add 475ml of H2O into the 500ml jar.
- Afterwards bring the jar to a laboratory steam sterilizer and put it in for 30 minutes.
- Finally pour the media into plates up to half way.
Heat Shock Procedure
- Calibration Experiment Procedure
- Set thermocycler to 48°C sitting temperature.
- Vortex the yeast culture briefly to resuspend the yeast cells.
- Label 6 pcr tubes A through F.
- Micropipette 90 ul of water into the pcr tubes.
- Micropipette 10 ul of each strain
- Label each tube “A, B, C, D, E, F”.
- Insert all 6 of the tubes into the thermocycler for exactly 42 minutes at 48°C.
- Afterwards remove all of the tubes from the thermocycler.
Dilution Procedure
- In a 96 channel well put in your six yeast strains then put them into the left most column desending from A to F.
- Then in the rest of the rows add 90 ul of H2O into A-F.
- After adding the water take 10 ul of the strains add them to the next column and mix.
Plating Procedure
- Label each petri dish accordingly, first plate "A,B", the Second plate "C,D", and the Third plate "E,F".
- Micropipette 5 ul of the strains onto the coordinated petri dish using the .
- Place all the petri dishes in the incubator (30°C).
- Let sit for 48 hours.
- Take out and analyze the growth.
- Repeat for 3 trials.
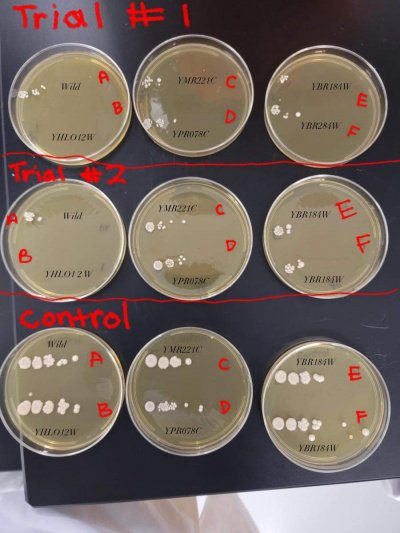
Results
- BY4735 (A)
- YHLO12W (B)
- YMR221C (C)
- YPRO78C (D)
- YBR184W (E)
- YBR284W (F)
Important Remaining Items (Remove Before Publish)
The raw data from the calibration experiment.
Your interpretation of the calibration experiment. Which parameter value did you settle on? Why?
[[Image:Heat Shock.jpeg|center|thumb|400px
[[Image:Calibration Experiment From Heat Shock.jpeg|center|thumb|400px
1). You repeated the same image on each gene page -- that's fine, but I can't tell which plate has which strain or condition on it! Which means that interpreting your data is basically impossible.
2). Additionally, you present an image that's the result of your calibration experiment -- but you never tell me what I'm looking at or what it means.
3). Finally, the details of the calibration protocol -- for how much time you shocked your cells -- seem wrong. Didn't you load them all in at the same time, then take them out at particular timepoints?
That's not what the protocol says. Also, please include the serial dilution that you did in your protocol!