Difference between revisions of "SGD Newsletter, Fall 2020"
Mjalexander (talk | contribs) (→Interaction Page Updates) |
Mjalexander (talk | contribs) (→Interaction Page Updates) |
||
| Line 15: | Line 15: | ||
==Interaction Page Updates== | ==Interaction Page Updates== | ||
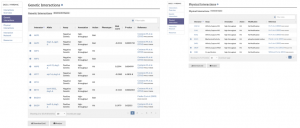
| − | [[File:interaction_page.png|link=https://yeastgenome.org/locus/S000001086/interaction|thumb| | + | [[File:interaction_page.png|link=https://yeastgenome.org/locus/S000001086/interaction|thumb|left|upright=1|Genetic interaction annotations (left) and Physical interaction annotations (right) in separate tables.]] |
SGD has made recent updates to our Gene Interactions Page for improved clarity. Previously, genetic and physical interaction annotations were combined in one table, but now these annotations are recorded in separate annotation tables. The menu in the top left corner can be used to view and navigate to each section of the Interactions page. Additionally, alleles, SGA Scores, and P-values are now included for annotations from the global interactions paper by Costanzo M, et al. (2016). | SGD has made recent updates to our Gene Interactions Page for improved clarity. Previously, genetic and physical interaction annotations were combined in one table, but now these annotations are recorded in separate annotation tables. The menu in the top left corner can be used to view and navigate to each section of the Interactions page. Additionally, alleles, SGA Scores, and P-values are now included for annotations from the global interactions paper by Costanzo M, et al. (2016). | ||
Revision as of 11:01, 2 December 2020
About this newsletter:
This is the Fall 2020 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can also subscribe to SGD's RSS feed to receive updates on SGD news:
Contents
New SGD Allele Pages
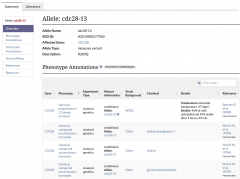
We are pleased to announce that SGD’s brand new Allele Pages are now available on our website. To navigate to an allele page, use the search bar to find a specific allele or enter a gene name and select an allele from the autocomplete list. Additionally, these pages can be accessed by clicking on the allele name in a gene’s Phenotype Annotation table. These pages are still being updated with new information as it becomes available.
The type of information that you can find on each allele page includes:
• Allele Overview: General information about the allele, such as its name, the affected gene, the type of allele (e.g. missense), and a description of sequence change and/or domain mutated.
• Phenotype and Interaction Annotations: Phenotype and Genetic Interaction Annotation tables for the allele.
• Shared Alleles: A network diagram depicting shared phenotypes and interactions with other alleles.
If you are interested in viewing all alleles for a specific gene or would like to view a comprehensive list of the alleles that SGD currently has curated, you can use this YeastMine template with your customized parameters.
Interaction Page Updates
SGD has made recent updates to our Gene Interactions Page for improved clarity. Previously, genetic and physical interaction annotations were combined in one table, but now these annotations are recorded in separate annotation tables. The menu in the top left corner can be used to view and navigate to each section of the Interactions page. Additionally, alleles, SGA Scores, and P-values are now included for annotations from the global interactions paper by Costanzo M, et al. (2016).
Research Spotlight (Blog Posts)
In case you missed them, here are some of the most popular Research Spotlights posted on our blog lately:
- Yeast on Their Best Behavior Just like you might change your behavior around friends and family this holiday season, Piccirillo and coworkers show that yeast also behaves differently because of the cells around them.
- Yeast, the Spam Filter Turns out yeast can act as a pretty good spam filter. Hamza and colleagues put it to the test when they systematically screened for human genes that can replace their yeast equivalents, and went on to test the function of tumor-specific variants in several selected genes that maintain chromosome stability in S. cerevisiae.
- Life Needs to be More Like a 1950’s Chevy Modern cars have a lot more bells and whistles than their older counterparts, but they just don’t seem to last as long! McCormick and coworkers showed a similar concept in their painstaking search through deletion strains were they found that many nonessential genes seem to shorten a yeast’s life.
- You Can Take Yeast Off of the Grapevine, But… Rossouw and colleagues show the importance of looking beyond lab isolation to get clues into the evolution of gene families.
- Unleashing the Awesome Power of Yeast Transcription In today’s age of social media, communicating across large distances is much easier than it used to be. Reavey and coworkers showed in a study that they can help yeast keep up with the times and mutate away it’s inability to activate transcription at a distance.
SGD at the 27th International Conference on Yeast Genetics and Molecular Biology (ICYGMB)

In September 2015 members of the SGD staff attended the 27th International Conference on Yeast Genetics and Molecular Biology in Levico Terme, Trento, Italy. Several of our members presented posters on various current SGD projects:
In addition to hosting an exhibit table where we got to interact with many of you, we were thrilled to have been given the opportunity to give an SGD workshop, Getting More Out of SGD, on the first day of the conference. The workshop featured exciting new data developments at SGD including new yeast-human functional complementation data, the new variant viewer, and new data in YeastMine. Your feedback is important to us, so we loved answering questions and hearing your comments at this well-attended workshop!
If you would like to see tweets from the conference, search #yeast2015 on Twitter.
Recent Publications from SGD Staff
- Sheppard TK, Hitz BC, Engel SR, Song G, Balakrishnan R, Binkley G, Costanzo MC, Dalusag KS, Demeter J, Hellerstedt ST, Karra K, Nash RS, Paskov KM, Skrzypek MS, Weng S, Wong ED, Cherry JM (2015). The Saccharomyces Genome Database Variant Viewer. Nucleic Acids Research. 2015 Nov 17; pii: gkv1250. doi: 10.1093/nar/gkv1250. PMID: 26578556
- Reid BJ, Culotti JG, Nash RS, Pringle JR (2015). Forty-five years of cell-cycle genetics. Mol Biol Cell. 2015 Dec 1;26(24):4307-12. doi: 10.1091/mbc.E14-10-1484. PMID: 26628751
What else have we been up to lately?
In addition to at the International Conference on Yeast Genetics and Molecular Biology, you may have seen SGD representatives at the following meetings:
- Senior biocuration scientist Edith Wong attended the annual ASCB meeting in December 2015.
- Senior biocuration scientists Marek Skrzypek and Maria Costanzo showcased SGD at the Cell Biology of Yeasts meeting in November at Cold Spring Harbor.
- Project Manager Gail Binkley attended the Genome Informatics meeting in October at Cold Spring Harbor.
- Senior biocuration scientist Rama Balakrishnan and PI Mike Cherry attended the GO Consortium meeting in Washington DC at the end of August.
Happy Holidays from SGD!
We would like to take this opportunity to recognize that 2020 has brought many changes and challenges for everyone. Our thoughts go out to all those who have been impacted by the unprecedented events of this year. We wish you and your family, friends, and lab mates the best during the upcoming holidays. Stanford University will be closed for three weeks starting on December 14 and will reopen on January 4th, 2021. Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will resume responding to user requests and questions in the new year.
Upcoming Meetings
- Colorado State University, Fort Collins CO, USA
- January 8, 2016
- Registration deadline extended: December 28, 2015
- Sant Feliu de Guixols, Spain
- June 11-16, 2016
- Lisbon, Portugal
- July 11-14, 2016
- Orlando, FL, USA
- July 13-17, 2016
- Abstract deadline: March 23, 2016
- Early registration deadline: March 23, 2016
- Awaji Yumebutai International Conference Center, Hyogo, Japan
- September 11-15, 2016