UW-Stout/Glucose SP23
Contents
Glucose Toxicity on BY4735 Strain: Pilot Experiment
Introduction
This study was provoked by a similar study into glucose sensitivity across different strains (Du & Lang, 2006). Once BY4735 strains’ reach a certain cell/ml count in glucose solution, they have been shown to initiate glucose-induced apoptosis for the survival of the colony as illustrated in the aforementioned study. To ensure replication of this study was feasible, a pilot experiment following said protocol was replicated. All solution transformations were conducted in a sterile environment for purity concerns. The controls were as follows: sterile water, 2% sorbitol (osmotic control), and 2% glucose. The hypothesis being tested was as follows; BY4735 will exhibit glucose-induced apoptosis after a 24-hour incubation period.
Materials/Equipment
- YPED rich medium plates (2% glucose, 2% peptone, 1% yeast extract)
- Autoclave
- BY4735 wild type strain
- Nanodrop(Spectrophotometer)
- 50 ml solutions of 2% glucose (m/v) and 2% sorbitol (m/v)
- 37°C shaking incubator
- 30°C incubator
- Centrifuge
- Biosafe hood
- 100ml flasks
Methods
- Bring BY4735 strain to stationary phase. Then, pellet in centrifuge and discard of excess liquid. Completing this washing phase 3 times.
- After third washing phase, resuspend in a small volume of water. Complete a 1/10 dilution cell culture nanodrop to determine a relative OD600.
- Cells are then transferred into 10ml of either 2% glucose, 2% sorbitol, or sterile water at 2x107 cells/ml final concentration.
- Incubate in 37°C shaking incubator at 150 rpm for 24 hours, plate and incubate in 30°C, then complete viability assay visually.
Results
The plates after incubation for 24 hours are pictured below (Figure 1).
Figure 1. YPD plates after 24 hour incubation period from top to bottom: 2% glucose, 2% sorbitol, sterile water.
Discussion
Plates provided inconclusive assay results and indicated that there was no difference between controls and therefore the hypothesis was rejected. Most likely, this was due to experimental error. One possible adjustment to make would be to stain and conduct a viability assay instead of plating. This will produce a more concise measurement of living versus dead cells to conduct a more accurate analysis of glucose-induced apoptosis within the population. This adjustment will be made when replicating the experiment on knockout strains. Ultimately, this experiment did not provide any scientific data. However, it helped prepare for the upcoming experiment.
References
Du, H., & Liang, Y. (2006). Saccharomyces cerevisiae ste20 mutant showing resistance to glucose-induced cell death. Yi chuan xue bao = Acta genetica Sinica , 33(7),
Glucose Toxicity on BY4735 & Knockout Strains
Introduction
This study was provoked by a similar study into glucose sensitivity across different strains (Du & Lang, 2006). Once BY4735 strains’ reach a certain cell/ml count in glucose solution, they have been shown to initiate glucose-induced apoptosis for the survival of the colony as illustrated in the aforementioned study. Wild type strain BY4735 was used as a control against the knockout strains. All solution transformations were conducted in a sterile environment for purity concerns. The controls were as follows: sterile water, 2% sorbitol (osmotic control), and 2% glucose. The hypothesis being tested was as follows; there will be significant glucose-induced apoptosis in every strain as compared to the control.
- Note** Additional measures were added regarding equipment, materials, and methods as compared to the pilot experiment. These measures were taken to reduce experimental error within our given lab environment. The protocol noted in the references was followed as closely as possible given these adjustments.
Materials/Equipment
- YPED rich medium (2% glucose, 2% peptone, 1% yeast extract)
- Autoclave
- Nanodrop (Spectrophotometer)
- 50ml solutions of 2% glucose (m/v) and 2% sorbitol (m/v)
- 37°C shaking incubator
- 30°C incubator
- Centrifuge
- Biosafe hood
- 100ml flasks
- Saccharomyces cerevisiae
- BY4735
- YMR144W
- SSK1
- YKL162C
- Trypan Blue (Cell dye)
- Microscope
- Hemocytometer
Methods
- Bring each strain to stationary phase in rich medium. Then, pellet in centrifuge and discard the excess liquid. Completing this washing phase 3 times. (Wash with sterile water)
- After the third washing phase, resuspend in a small volume of water. Complete a 1/10 dilution cell culture nanodrop to determine a relative OD600.
- Cells are then transferred into 10ml of either 2% glucose, 2% sorbitol, or sterile water at 2x107 cells/ml final concentration.
- Incubate in 37°C shaking incubator at 150 rpm for 24 hours.
- Mix 1 part cells and 1 part trypan blue.
- Count number of cell deaths using hemocytometer.
Results
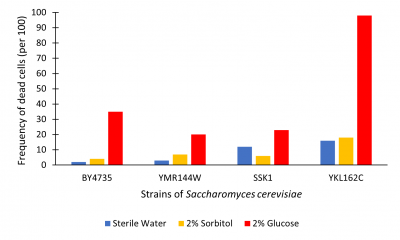
The graph below illustrates cell death as read by the hemocytometer as number of dead cells/100.
Figure 2. Viability assay from hemocytometer; measured dead cell per 100.
Discussion
Results were determined by viewing cell death (See Figure 2). Death was determined using trypan blue. Cells that were dead become died blue because their membrane no longer hold out the dye. Each trial we counted the number of cells to approximately 100-200 to cells that lived vs died. This was to approximate the percent of cells that had died. From what we found, in sterile water nearly all cells lived across the test. YKL162C gene seemed to have higher levels of death throughout all trials. 2% sorbitol and sterile water on average had little effect on cell growth the outlier being SSK1 where sterile water was about 5% higher. 2% glucose had higher levels of death in all trials. In the YKL162C 2% glucose trial nearly all cells had died. Our findings were that cells incubated for 48 hours in 2% glucose have increased cell death; most likely glucose-induced apoptosis as illustrated in the BY4735 population.
The YKL162C gene may have some effect on cell membrane efficiency because of the increased death across the board and for complete death in glucose trial. Compared to BY4735, this knockout seemed to produce extreme glucose-induced apoptosis. So, in removing this gene it may decreases cell survivability as whole; further research into this is suggested.
References
Du, H., & Liang, Y. (2006). Saccharomyces cerevisiae ste20 mutant showing resistance to glucose-induced cell death. Yi chuan xue bao = Acta genetica Sinica , 33(7),