UW-Stout/Formamide SP22
Contents
Wild Yeast Cell Pilot Procedure
Caution
Neat Formamide is harmful to the eyes, if swallowed, inhaled, or absorbed through the skin.
Materials / Equipment
- Neat Formamide, 33ul
- Sterile water, 567ul
- 0.2ml flat cap PCR tube, 13 tubes
- Corning COSTAR 96-well clear flat-bottom assay plate
- Wild-type yeast in 2x synthetic complete media at an OD600 of 0.1-0.2, 600ul
- Molecular Devices SpectraMax Plus 384 Microplate Reader
- Micropipet (100ul)
- Micropipet (10ul)
- Micropipet (2ul)
- Micropipet tips
Formamide Concentrations
- 0.5ul formamide + 49.5ul sterile water
- 1.0ul formamide + 49.0ul sterile water
- 1.5ul formamide + 48.5ul sterile water
- 2.0ul formamide + 48.0ul sterile water
- 2.5ul formamide + 47.5ul sterile water
- 3.0ul formamide + 47.0ul sterile water
- 3.5ul formamide + 46.5ul sterile water
- 4.0ul formamide + 46.0ul sterile water
- 4.5ul formamide + 45.5ul sterile water
- 5.0ul formamide + 45.0ul sterile water
- 5.5ul formamide + 44.5ul sterile water
- 0.0ul formamide + 50.0ul sterile water
Procedure
- Obtain the neat formamide (refrigerated) and sterile water
- Obtain and label 12 PCR tubes 1-12
- Obtain one more PCR tube to transfer 33ul of neat formamide inside, can put excess formamide in
- Using the 13th PCR tube, pipet appropriate amount of formamide and sterile water into the labeled PCR tubes according to number
- In a sterile environment, pipet the solution (50ul) from the PCR tube into the well cell
- Vortex the yeast culture briefly to resuspend the yeast cells, and then pipet 50ul of the wild yeast cells into each well in addition to the formamide-water solution
- Set up the plate reader as follows:
- Temperature: 30 degrees Celsius
- Mode: kinetic
- Wavelength: 600 nm
- Interval: 5 minutes
- Total run time: 24 hours
- Shake before read: 30 seconds
- Transfer the assay plate to the reader and read for 24 hours
- Record data
Results
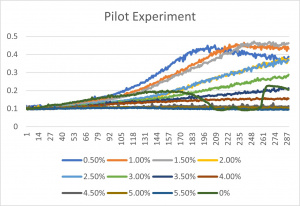
- The x-axis is in time (min) and the y-axis is growth by concentration
- The x-axis corresponds to the concentrations listed up above under the title "formamide concentrations"
- 0% is an experimental error- since it has a concentration of 0% formamide anyway, it was left in the final graph
- The x-axis is in time (min) and the y-axis is growth by concentration
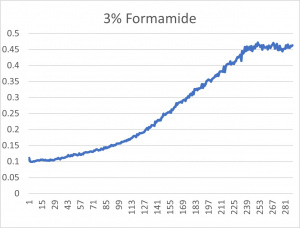
- The 3% formamide used in the pilot experiment and represented as the light blue color (in the top-most picture) was chosen to be used on the transformed yeast cells
- This was deduced due to the position of the yeast cells under 3% formamide stress
- Under 3% formamide, the yeast cells were able to grow, but didn't grow too much- simply put, the 3% line was right in the middle of the graph
Knockout Yeast Cell Gene
Materials / Equipment
- Neat formamide, 18ul
- Sterile water, 423ul
- 0.2ml flat cap PCR tube, 10 tubes
- Well plate
- Knock out yeast cells
- Molecular Devices SpectraMax Plus 384 Microplate Reader
- Micropipet (100ul)
- Micropipet (20ul)
- Micropipet (2ul)
- Micropipet tips
Procedure
- Obtain neat formamide (refrigerated) and sterile water
- Obtain and label PCR tubes 1-9
- Pipet approximately 18ul formamide into a 10th PCR tube, can put excess formamide in
- Using the 10th PCR tube, pipet 3.0ul of formamide into each PCR tube (labeled 1-9)
- Pipet 47.0ul of sterile water into each of the numbered PCR tubes
- In a sterile environment, pipet 50ul of formamide-water solution from the PCR tubes into well cells
- Vortex each yeast culture strain briefly to resuspend the cells, and then pipet 50ul of each knockout strain into the appropriate well cells in addition to the solution
- Set up the plate reader as follows:
- Temperature: 30 degrees Celsius
- Mode: kinetic
- Wavelength: 600 nm
- Interval: 5 minutes
- Total run time: 24 hours
- Shake before read: 30 seconds
- Transfer the assay plate to the reader and read for 24 hours
- Record data
Results
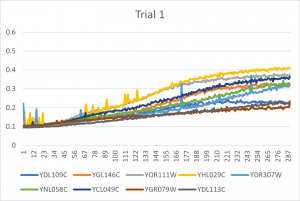
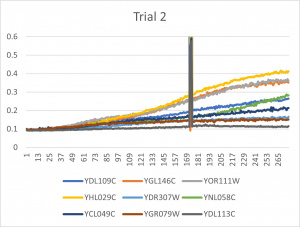
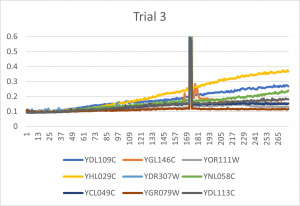
- Ran three trials, trial one through three is posted up above in picture form as a line graph
- The dark line in the middle of trail 2 and 3 is a scanning error from the Molecular Devices SpectraMax Plus 384 Microplate Reader
- Computed doubling times from the average times of the three trials for further analyzation