SGD Newsletter, Spring 2021
About this newsletter:
This is the Summer 2021 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community.
You can also subscribe to SGD's RSS feed to receive updates on SGD news:
http://www.yeastgenome.org/feed
Contents
R64.3 Annotation Update
SGD curators periodically update the chromosomal annotations of the S. cerevisiae Reference Genome, which is derived from strain S288C.
The R64.3 annotation release included various updates and additions:
- 7 new ORFs: OTO1/YGR227C-A, YHR052C-B, YHR054C-B, YJR107C-A, YKL104W-A, YLR379W-A, YMR008C-A
- 5 new ncRNAs: GAL10-ncRNA, TBRT/XUT_2F-154, SUT169, PHO84 lncRNA, GAL4 lncRNA
- 2 new uORFs for ROK1/YGL171W
- 1 new recombination enhancer: RE
- 1 new LTR: YELWdelta27
- 3 ORFs with shifted translation starts: HPA3/YEL066W, YJR012C, LTO1/YNL260C
- 1 ORF with shifted translation stop plus new intron: LDO45/YMR147W
- Changed feature_type (and SO_term) for non-transcribed spacers: NTS1-2, NTS2-1, NTS2-2
- New systematic nomenclature system for entire annotated complement of ncRNAs
Various sequence and annotation files are available on SGD's Downloads site. You can find more update details and see the new systematic nomenclature system for noncoding RNA genes on the Details of 2021 Reference Genome Annotation Update R64.3 SGD Wiki page.
Alliance Item
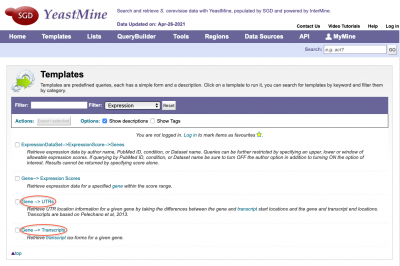
YeastMine Updates and New Templates
SGD has updated the current Gene-->UTRs YeastMine template with newly calculated 5' and 3' UTR sequence/coordinates. Additionally, Transcript iso-forms for specific genes from the Pelachano et al 2013 study can be accessed in YeastMine using the new Gene-->Transcript template. Both templates can be found under the 'Templates' section of YeastMine under the 'Expression' category.
Textpresso Update
Textpresso has recently been updated with a new system, bringing an overhauled user interface and new features. The updated Textpresso still allows users to search the corpus of texts with specialized queries using keywords and related categories, but now search results are shown in the context of the full text. More information about the changes and types of papers stored in Textpresso can be found in their About Us help section.
Fungal Pathogen Genomics Workshop
From May 10th - 14th, Senior Biocuration Scientist Edith Wong, Senior Biocuration Scientist Rob Nash, Senior Biocuration Scientist Marek Skrzypek, Biocuration Scientist Suzi Aleksander, and Associate Biocuration Scientist Micheal Alexander attended the Fungal Pathogen Genomics Workshop. Our curators helped attendees learn more about using the unique tools on our website and were also able to interact with curators from other fungal databases at FungiDB, EnsemblFungi, PomBase, CGD, MycoCosm, and JGI.
We would like to thank the Fungal Pathogen Genomics team for hosting a fun and successful virtual workshop for experimental biologists to gain valuable experience!
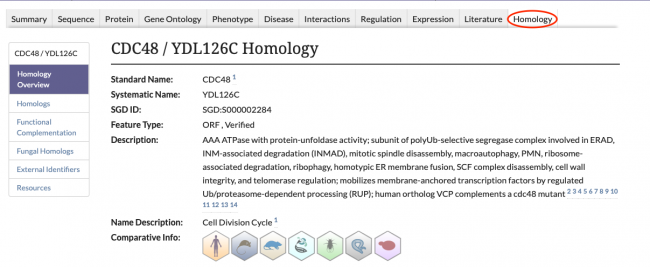
New Homology Pages
SGD is excited to introduce our new Homology Pages! These pages can be accessed by clicking on the Homology tab in the header of SGD gene pages, as seen below.
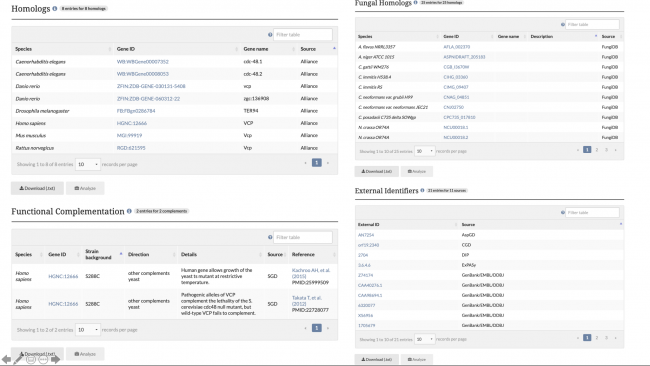
The information displayed on the Homology Pages is divided into several sections:
- Homologs: Information about known homologs for the gene of interest, such as the species of the homolog, the corresponding Gene ID from the Alliance of Genome Resources, and the name of the homolog.
- Functional Complementation: Data about cross-species functional complementation between yeast and other species, curated by SGD and the Princeton Protein Orthology Database (P-POD).
- Fungal Homologs: Curated homolog information for 24 additional species of fungi. View the species of the fungal homolog, the database source of the entry, and the Gene ID of the homolog from that database.
- External Identifiers: A list of external identifiers for the protein from various database sources.
Functional Complementation Data Available on References Pages
Functional Complementation annotations are now viewable on reference pages for which there is curatable functional complementation data. This information describes cross-species functional complementation between yeast and other species, and is curated by SGD and the Princeton Protein Orthology Database (P-POD).
insert picture
More Alleles
SGD now has approximately 13,000 alleles that are either fully or partially curated. You can generate a list of all alleles in our database using the Genes --> Alleles template in YeastMine, or search for a specific allele in the website search bar.