YHR048W
Share your knowledge...Edit this entry! <protect>
| Systematic name | YHR048W |
| Gene name | YHK8 |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr VIII:204607..206151 |
| Primary SGDID | S000001090 |
Description of YHR048W: Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles[1][2]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
DNA and RNA Details
Other DNA and RNA Details
Other Topic: expression
Specifically lower expression in carbon limited chemostat cultures versus carbon excess. [3] [4]
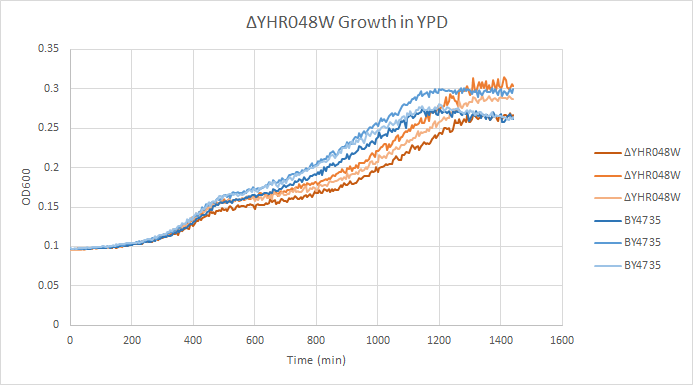
Growth in YPD
In a BY4735 background, knocking out YHR048 seems to have little to no effect on growth rate in log-phase. In this assay, the BY4735 strain's doubling time was 414 minutes, while the YHR048W knock-out strain's doubling time was 432 minutes. (These doubling times are the means of three experiments.)
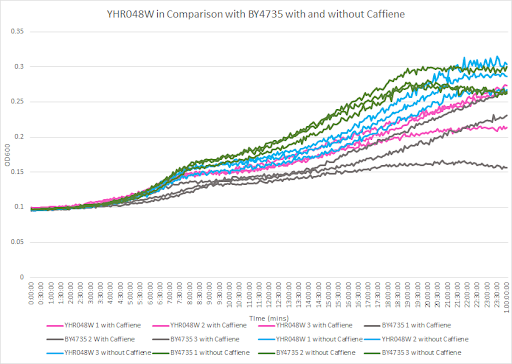
Caffeine and Yeast Cells
This graph shows the growth curve with and without caffeine. It has the knockout strain and wild type yeast cells. The caffeine enhanced the growth in both wild type and knockout strain. The average doubling time between the three experiments for the knockout strand was 801.97 minutes. The caffeine also had a positive effect on the growth curve for the wild type with the average doubling time being 1245.02 minutes. The results were concluded from the wild type and knockout strand was with this amount of caffeine it leads to an increase in growth.
The protocol can be found at
Caffeine
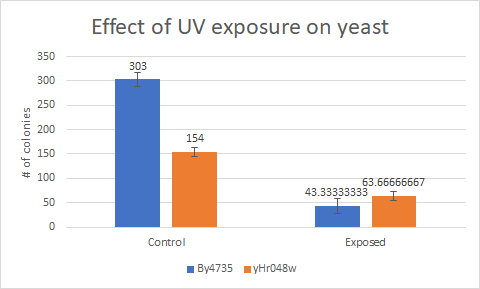
This bar graph shows that compared to a BY4735 background, Knocking out yHR048c makes yeast less sensitive to UV light exposure
The protocol can be found at
UW_Stout/UV_Light_FA19UV Light Exposure
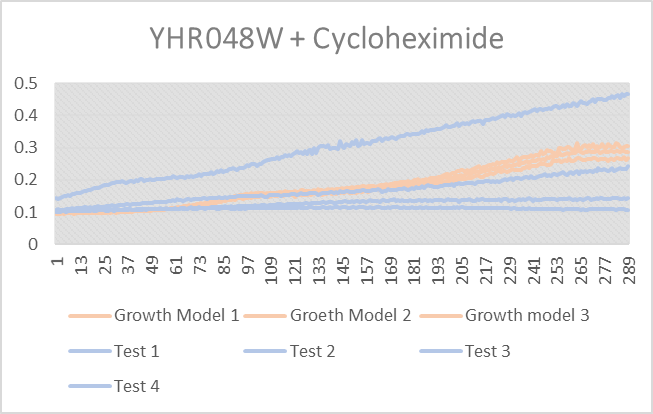
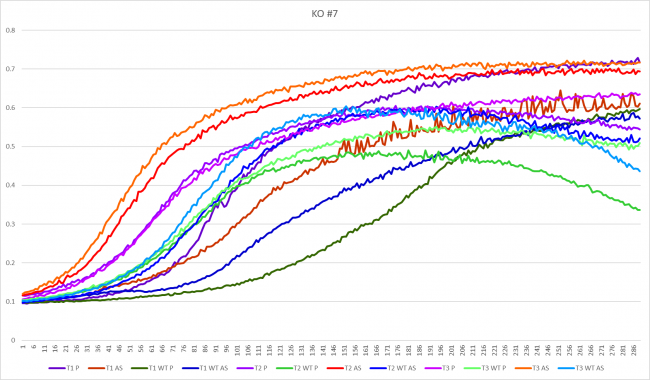
Cycloheximide effect on YHR048W
As you can see from above test 4 thrived, it did very good, but test 1-3 did not do as well and were all lower than the growth curve. Growth models 1-3 are YHR048W without any stress induced.
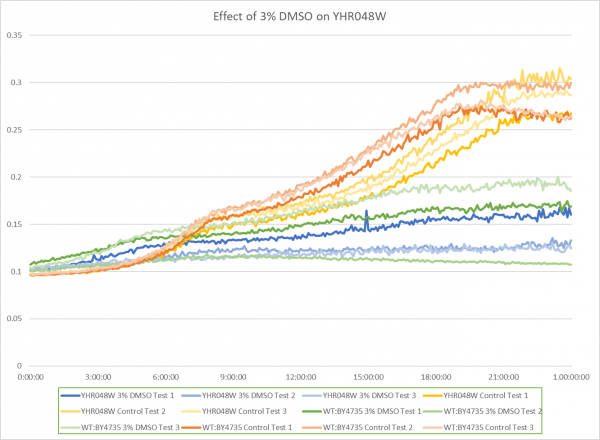
Effect of 3% DMSO on YHR048W
The graph above shows the effect of 3% DMSO on YHR048W. From the data represented the average doubling rate for the YHR048W knockout strain without DMSO was 575 minutes. When DMSO is added the average doubling time increased to 1851 minutes. This shows that 3% DMSO did add stress to this knockout strain. The average doubling time of WT:BY4735 without DMSO was 514 minutes. The average doubling rate for WT:BY4735 with DMSO was 1529 minutes. If we compare the average doubling rate of WT:BY4735 with DMSO to YHR048W with DMSO we can see that without the YHR048W gene the yeast was unable to grow as quickly. This suggest that the YHR048W gene does play a role in protecting yeast against oxidative stress induced by DMSO. If we compare the growth rates of the controls for both WT:BY4735 and YHR048W we can see that the YHR048W gene barely effects the growth of yeast in optimal conditions.
Growth in Proline and Ammonium Sulfate
In YHR048W, this strand seemed to grow better in the proline media with an average doubling time of 36.27 minutes. The average doubling time in ammonium sulfate was 40.47 minutes. This was opposite to the wild types though. The wild type strand grew faster in ammonium sulfate at an average doubling rate of 46.03 minutes. When grown in proline, the average doubling time was not much slower, with a time of 48 minutes.
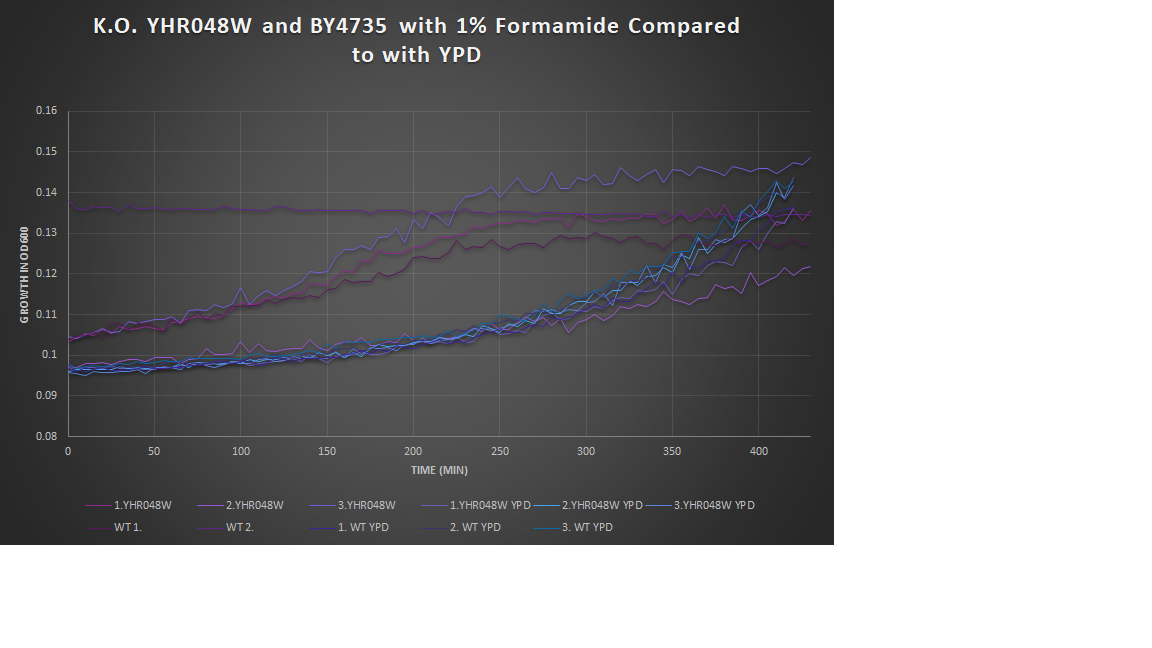
1% Formamide Yeast Torture[[1]]
This graph shows the growth curve of the knockout yeast strain YHR048W and wild type BY4735 in 1% formamide in 3 trials. The average calculated doubling time for YHR048W with 1% formamide was 2,546.13 minutes, while the unstressed strain was 2,698.96 minutes. This concluded that the stressed out strain is not too sensitive to 1% formamide, and grows at a faster pace than the unstressed strand in this environment. Our stressed wild types doubling time was 2,865.53 minutes, and the unstressed was 3,268.13 minutes. The stressed wild type in this experiment proved to show better growth concluding that the addition of 1% formamide for this strain does not cause sensitivity.
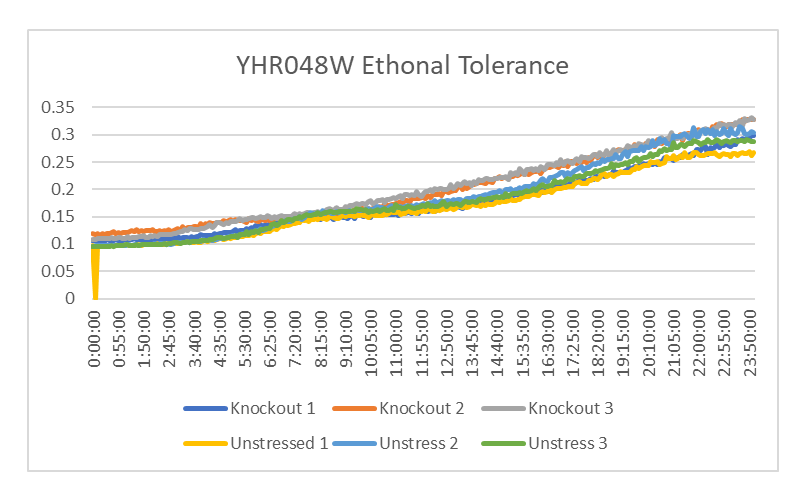
Ethanol Tolerance in YHR048W
This graph shows the growth between YHR048W treated and untreated with ethanol. The rate of growth of the yeast treated with ethanol is 1680.033 minutes. The rate of growth of the yeast not treated with ethanol is 1638.69 minutes. This shows that the strands of YHR048W treated with ethanol have a similar growth rate than the ones that weren't.
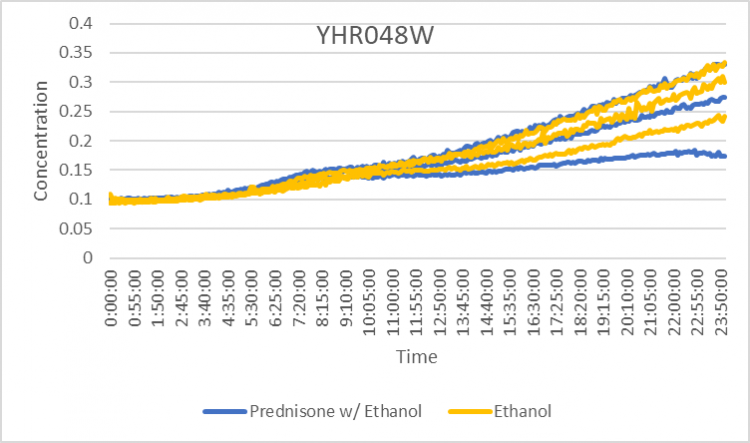
Prednisone Effects on YHR048W
The graph above shows how dilutions P2 (30 µg/ml) and E2 (10% ethanol solution) impacted the growth rate of the knockout yeast strain YHR048W over 24 hours. The concentration did not double for test 3 of P2 for this experiment, so by taking only the first two test results, the doubling rate averages to around 1,020 minutes for only ethanol. As for the Prednisone with ethanol, the doubling rate averages to around 925 minutes. Overall, the Prednisone increased the rate of growth by almost 95 minutes. These results conclude that Prednisone promotes the growth of yeast cells without the YHR048W gene.
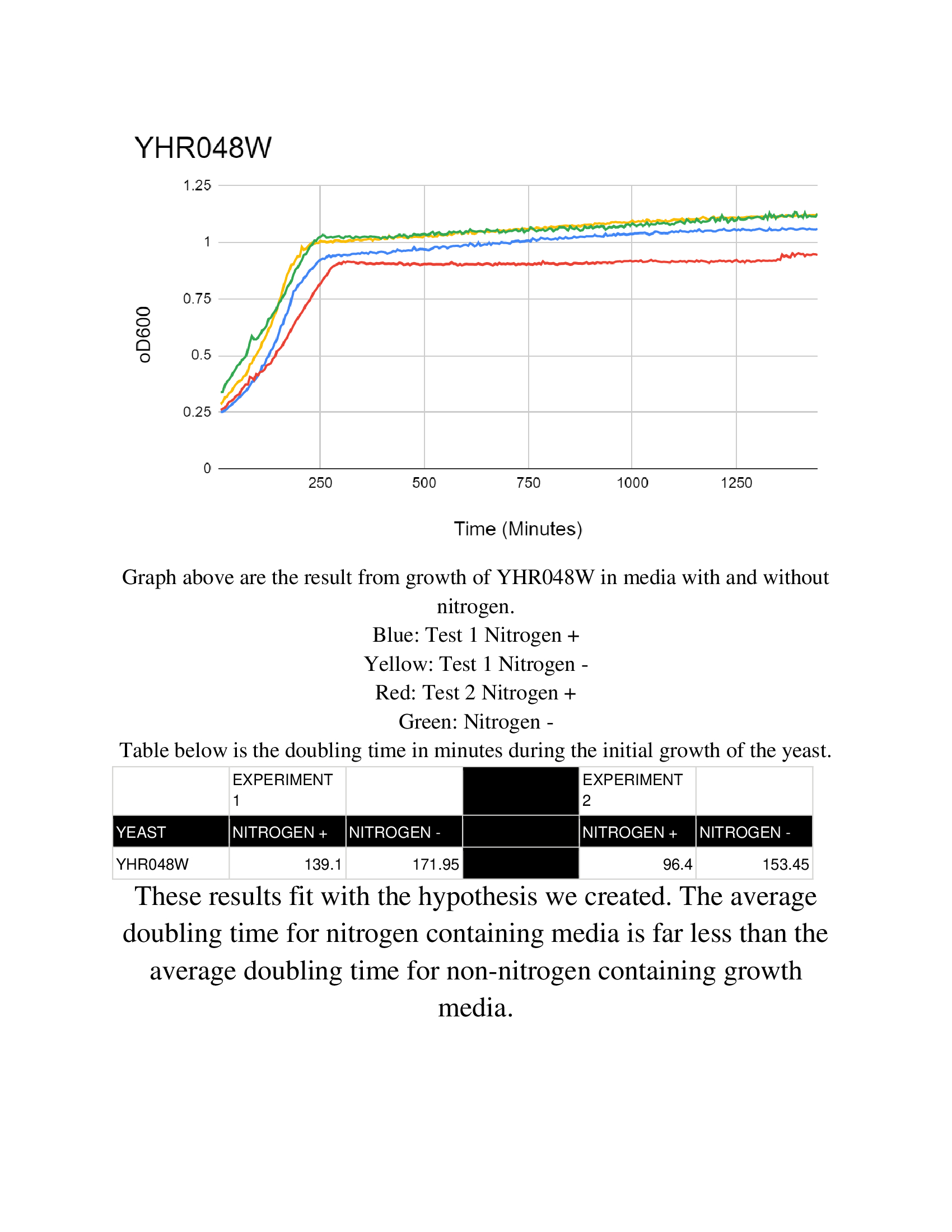
The Wild Type BY4735 has data located at the Nitrogen Starvation Protocol.
Nitrogen starvation
Hydroxyurea
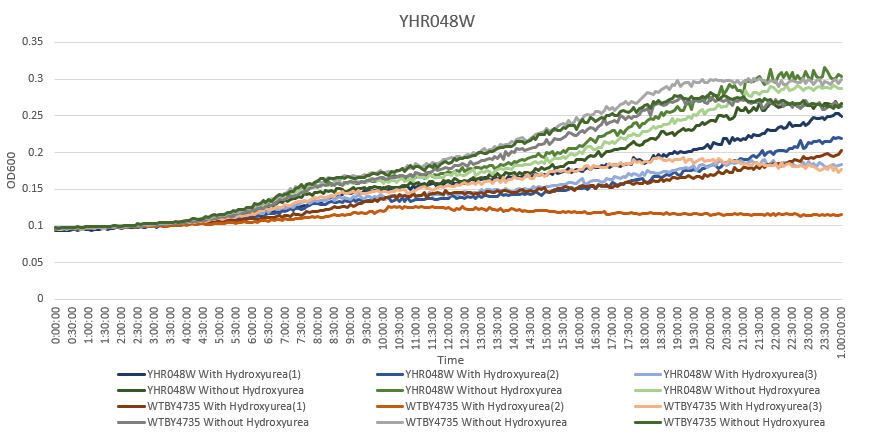 The averaging doubling time for YHR048W is 572.25 minutes. The averaging doubling time for the unstressed YHR048W is 365.74 minutes
The averaging doubling time for YHR048W is 572.25 minutes. The averaging doubling time for the unstressed YHR048W is 365.74 minutes
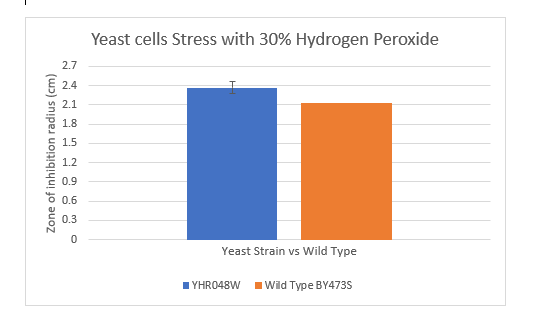
Zone of Inhibition of 30% Hydrogen Peroxide on YHR048W
References
See Help:References on how to add references
- ↑ Barker KS, et al. (2003) Identification of genes differentially expressed in association with reduced azole susceptibility in Saccharomyces cerevisiae. J Antimicrob Chemother 51(5):1131-40 SGD PMID 12697649
- ↑ Gbelska Y, et al. (2006) Evolution of gene families: the multidrug resistance transporter genes in five related yeast species. FEMS Yeast Res 6(3):345-55 SGD PMID 16630275
- ↑ Boer VM, et al. (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278(5):3265-74 SGD PMID 12414795
- ↑ submitted by Viktor Boer on 2003-07-25
See Help:Categories on how to add the wiki page for this gene to a Category </protect>