Difference between revisions of "YLR006C"
(→Acids) |
(→Acids) |
||
| Line 42: | Line 42: | ||
[[UW-Stout/Acids_SP23]] | [[UW-Stout/Acids_SP23]] | ||
| − | [[File:MEL_control_v.s_knockout_ | + | [[File:MEL_control_v.s_knockout_#3]] |
==Caffeine== | ==Caffeine== | ||
Revision as of 18:57, 2 May 2023
Share your knowledge...Edit this entry! <protect>
| Systematic name | YLR006C |
| Gene name | SSK1 |
| Aliases | |
| Feature type | ORF, Verified |
| Coordinates | Chr XII:163893..161755 |
| Primary SGDID | S000003996 |
Description of YLR006C: Cytoplasmic response regulator; part of a two-component signal transducer that mediates osmosensing via a phosphorelay mechanism; required for mitophagy; dephosphorylated form is degraded by the ubiquitin-proteasome system; potential Cdc28p substrate[1][2][3][4]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
Protein Details
Protein Modification
Modification(s): Phosphorylation
Identified as an efficient substrate of Clb2-Cdk1-as1 in a screen of a proteomic GST-fusion library. [4] [5]
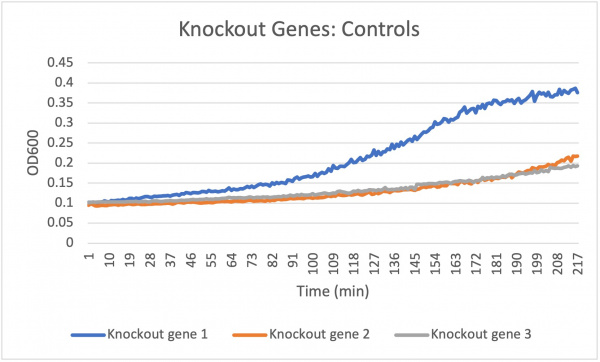
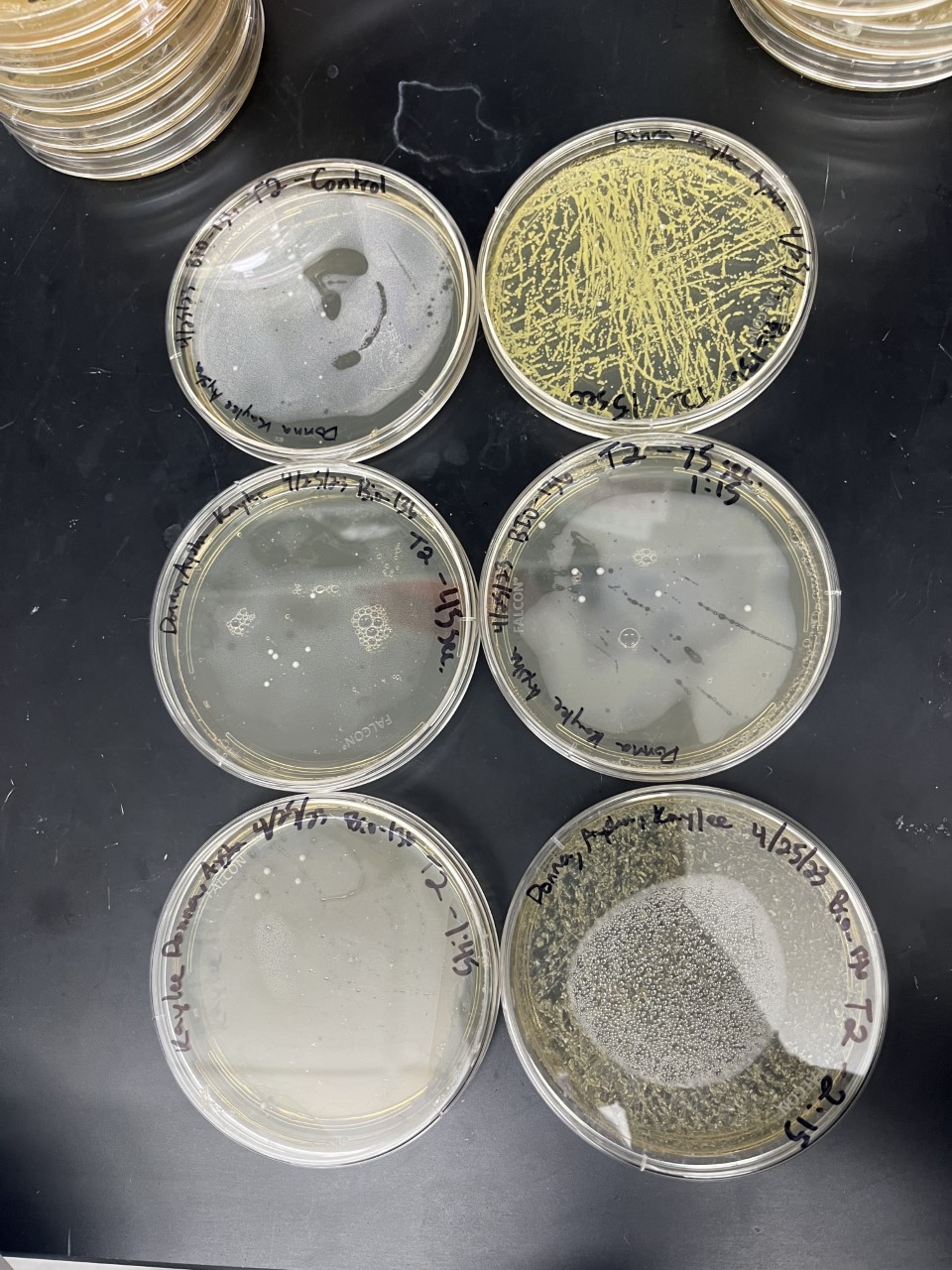
Acids
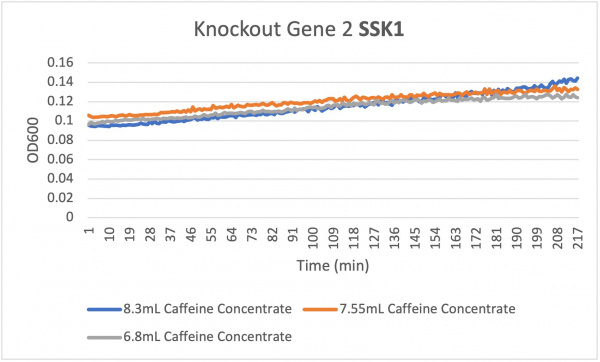
Caffeine
Analysis
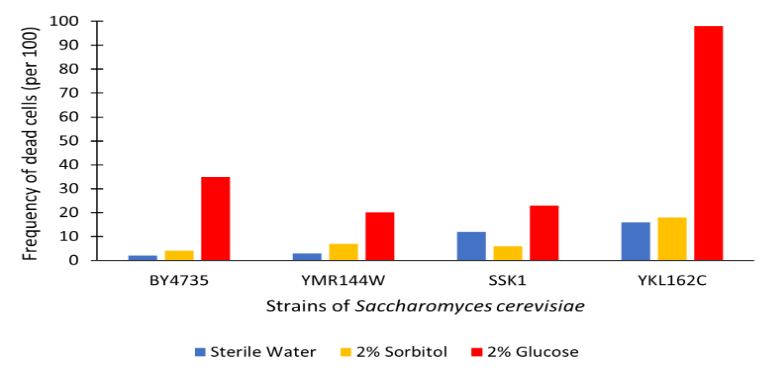
Glucose Sensitivity
Protocol
Analysis
The graph displayed above shows a viability assay of SSK1 knockout gene as compared to BY4735 (wild type) and other knockout strains. We found that there was about 20% less cell death then in the control wild type. It was surprising to find that by removing this gene there was an increase in cell viability. Further research may be conducted to test this slight resistance to all cell death, but more importantly the resistance to glucose-induced apoptosis as noted in the wild type BY4735 for more conclusive results.
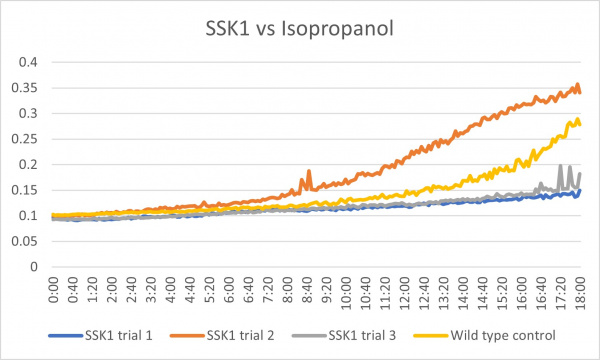
Isopropanol
Analysis
These results were what was expected with only a little bit of difference. The first trial and third trials were close and there was less growth than the control which could have been because the knockout genes were more sensitive to the isopropanol. The second trial gave different results in which there was more growth than the control which is the opposite of the first and third. This could have been caused by an error because there was consistency in the other two.
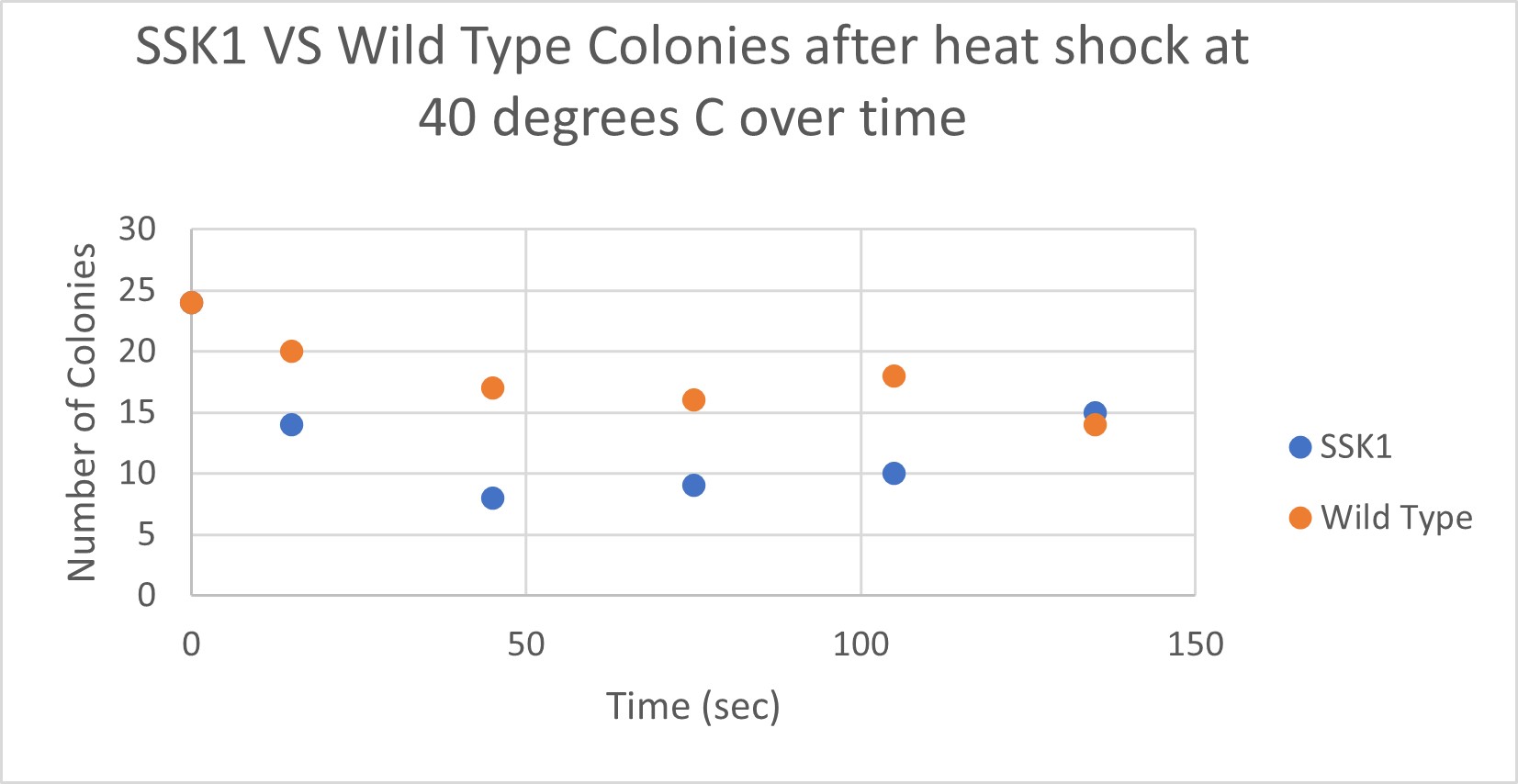
Heat Shock
| Time (sec) | Colonies | |
|---|---|---|
| 0 | 24 | |
| 15 | 14 | |
| 45 | 8 | |
| 75 | 9 | |
| 105 | 10 | |
| 135 | 15 |
| Time (sec) | Colonies | |
|---|---|---|
| 0 | 24 | |
| 15 | 20 | |
| 45 | 17 | |
| 75 | 16 | |
| 105 | 18 | |
| 135 | 14 |
Analysis
The general trend of our data yielded a negative relationship between time of heat shock. About half of the yeast cells died off in a matter of 135 seconds at 40 degrees Celsius. The SSK1 yeast were dying off at a very comparable rate over time compared to the Wild Yeast strain. This means that the gene that was knocked out must not have a correlation with heat shock.
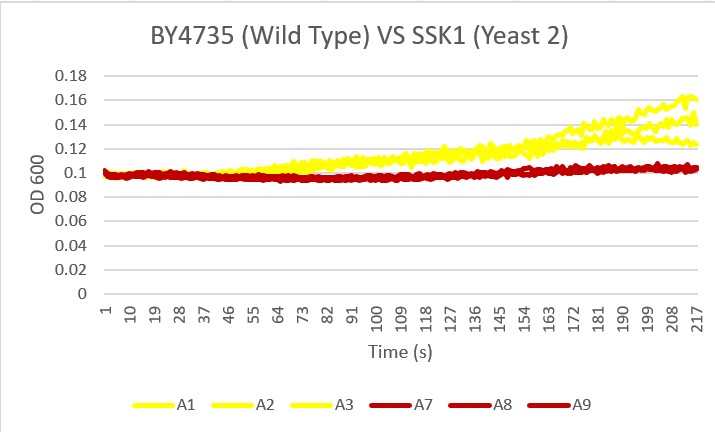
Bases
Analysis Yeast 2, A4 A5 A6, showed almost no growth compared to the Wild Type yeast, A1 A2 A3. This knockout gene likely did not benefit the resistance to basic pH changes in the environment.
Protocol- UW-Stout/Bases SP23
References
See Help:References on how to add references
- ↑ Mao K, et al. (2011) Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol 193(4):755-67 SGD PMID 21576396
- ↑ Posas F, et al. (1996) Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 two-component osmosensor. Cell 86(6):865-75 SGD PMID 8808622
- ↑ Sato N, et al. (2003) Phosphorelay-regulated degradation of the yeast Ssk1p response regulator by the ubiquitin-proteasome system. Mol Cell Biol 23(18):6662-71 SGD PMID 12944490
- ↑ 4.0 4.1 Ubersax JA, et al. (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425(6960):859-64
SGD PMID 14574415 Cite error: Invalid
<ref>tag; name "S000074306" defined multiple times with different content - ↑ submitted by Jeff Ubersax on 2004-01-29
See Help:Categories on how to add the wiki page for this gene to a Category </protect>