Difference between revisions of "YKL162C"
(→Bases) |
(→Bases) |
||
| Line 105: | Line 105: | ||
[[Image:EMYeast3.jpg]] | [[Image:EMYeast3.jpg]] | ||
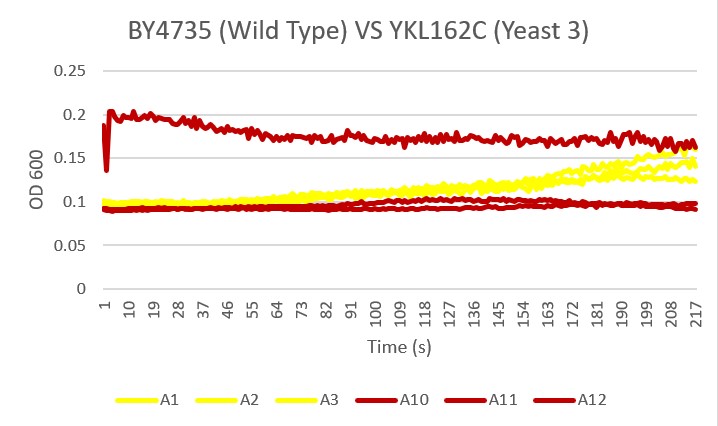
| − | '''Analysis''' As you can see there was clearly an error with the A11 sample of yeast 3. The other two samples showed slight growth over the 24 hour time period. They did not seem to be bothered by the ammonium chloride and ammonium hydroxide buffer. | + | '''Analysis''' |
| + | As you can see there was clearly an error with the A11 sample of yeast 3. The other two samples showed slight growth over the 24 hour time period. They did not seem to be bothered by the ammonium chloride and ammonium hydroxide buffer. | ||
==References== | ==References== | ||
Revision as of 13:01, 2 May 2023
Share your knowledge...Edit this entry! <protect>
| Systematic name | YKL162C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr XI:148838..147630 |
| Primary SGDID | S000001645 |
Description of YKL162C: Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the mitochondrion[1]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
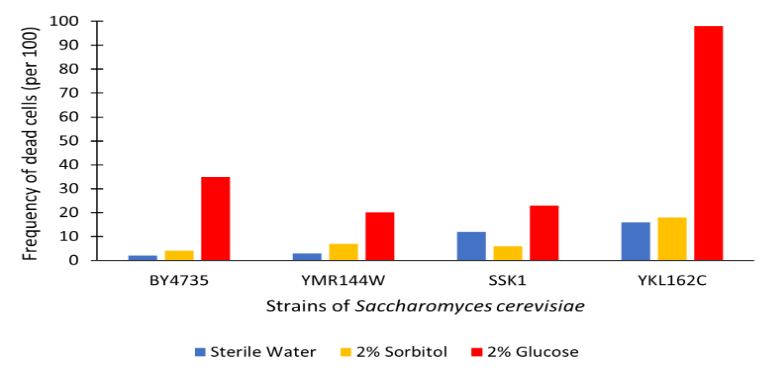
Glucose Sensitivity
[1]Protocol
Analysis
The table above shows the number of cells with YKL162C removed that were counted as dead out of 100 counted cells. We found that in all trials compared to the control there was in increase in cell death. With the YKL162C gene removed nearly all cells died when incubated for 48 hours in 2% Glucose. This knockout may intrigue further research into basic cell viability as a result of the statistical significance.
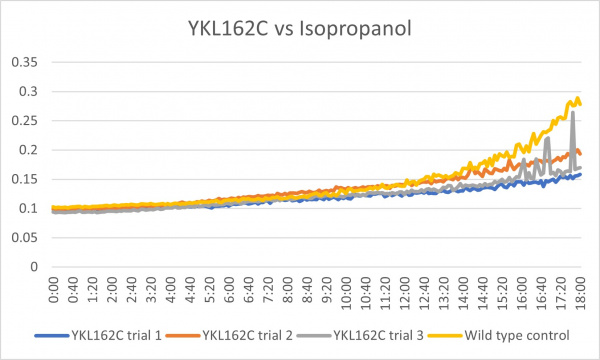
Isopropanol
Analysis
These results were what was expected and all 3 were consistent throughout the trials. Although there was less growth than the control group, it could mean that the cell is more sensitive to the isopropanol with the gene knocked out. In the third trial there was a little bit of spiking towards the end around 15:20-18:00 but other than that it was a good growth curve.
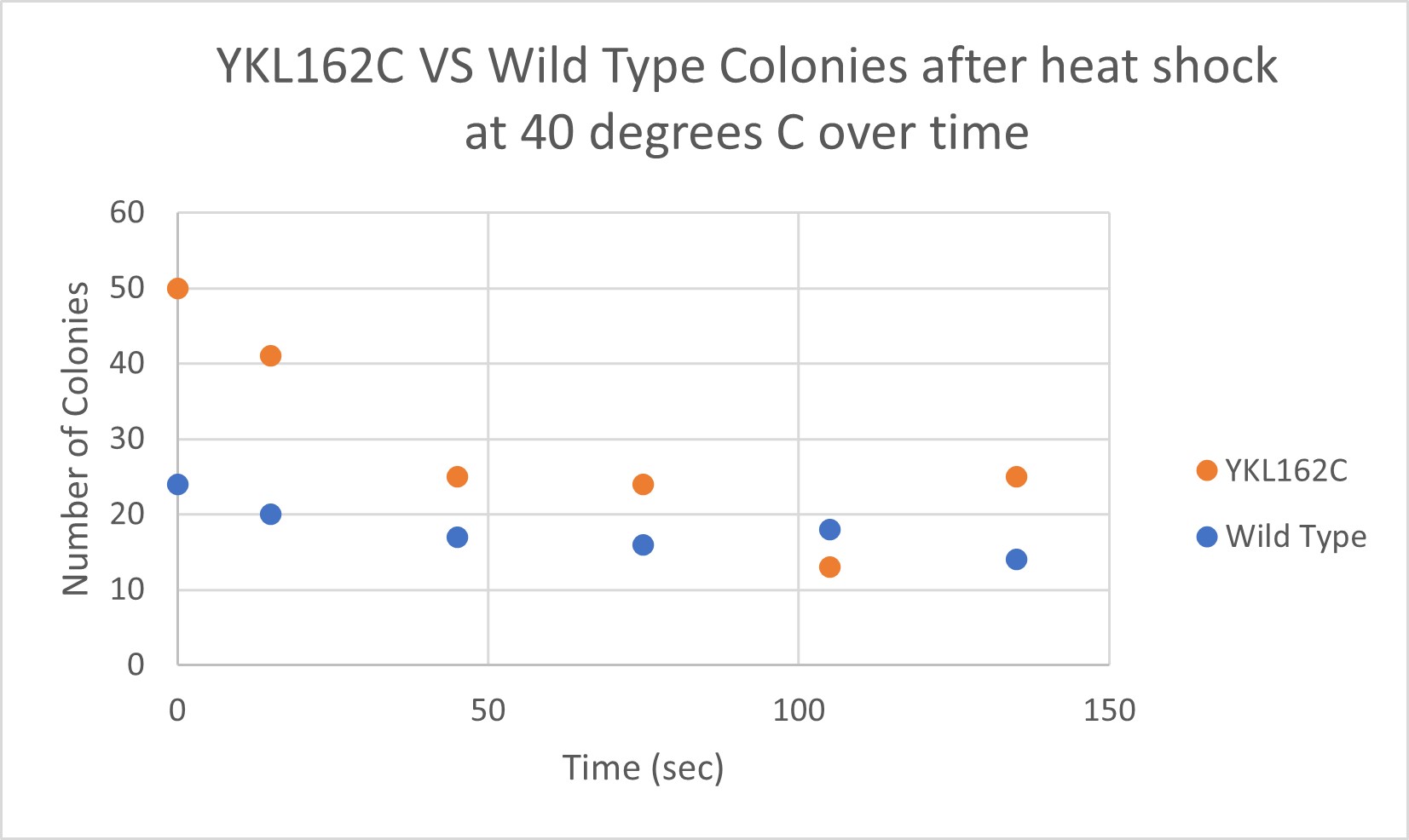
Heat Shock
| Time (sec) | Colonies | |
|---|---|---|
| 0 | 50 | |
| 15 | 41 | |
| 45 | 25 | |
| 75 | 24 | |
| 105 | 13 | |
| 135 | 25 |
| Time (sec) | Colonies | |
|---|---|---|
| 0 | 24 | |
| 15 | 20 | |
| 45 | 17 | |
| 75 | 16 | |
| 105 | 18 | |
| 135 | 14 |
Analysis
The general trend of our data yielded a negative relationship between time of heat shock. About half of the yeast cells died off in a matter of 135 seconds at 40 degrees Celsius. The YKL162C yeast were dying off at a faster rate over time compared to the Wild Yeast strain. This means that the gene that was knocked out in the YKL162C yeast could have cause some resistance to heat shock.
Bases
Analysis As you can see there was clearly an error with the A11 sample of yeast 3. The other two samples showed slight growth over the 24 hour time period. They did not seem to be bothered by the ammonium chloride and ammonium hydroxide buffer.
References
See Help:References on how to add references
See Help:Categories on how to add the wiki page for this gene to a Category </protect>