Difference between revisions of "YBR033W"
| Line 35: | Line 35: | ||
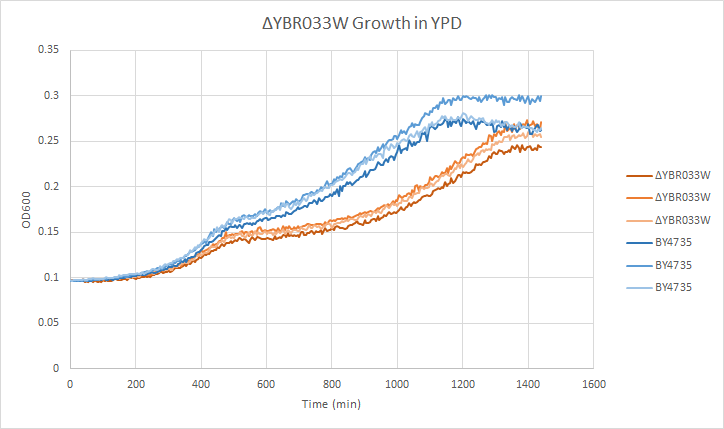
In a BY4735 background, knocking out YBR033W seems to have a moderate effect on growth rate in log-phase. In this assay, the BY4735 strain's doubling time was 414 minutes, while the YBR033W knock-out strain's doubling time was 532 minutes. (These doubling times are the means of three experiments.) | In a BY4735 background, knocking out YBR033W seems to have a moderate effect on growth rate in log-phase. In this assay, the BY4735 strain's doubling time was 414 minutes, while the YBR033W knock-out strain's doubling time was 532 minutes. (These doubling times are the means of three experiments.) | ||
| + | |||
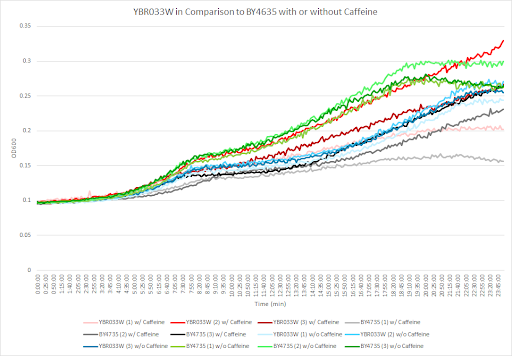
Caffeine and Yeast Cells | Caffeine and Yeast Cells | ||
Revision as of 06:38, 12 December 2019
Share your knowledge...Edit this entry! <protect>
| Systematic name | YBR033W |
| Gene name | EDS1 |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr II:301944..304703 |
| Primary SGDID | S000000237 |
Description of YBR033W: Putative zinc cluster protein, predicted to be a transcription factor; YBR033W is not an essential gene[1][2]
</protect>
Community Commentary
About Community Commentary. Please share your knowledge!
Growth in YPD
In a BY4735 background, knocking out YBR033W seems to have a moderate effect on growth rate in log-phase. In this assay, the BY4735 strain's doubling time was 414 minutes, while the YBR033W knock-out strain's doubling time was 532 minutes. (These doubling times are the means of three experiments.)
Caffeine and Yeast Cells
<protect>
References
See Help:References on how to add references
- ↑ MacPherson S, et al. (2006) A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70(3):583-604 SGD PMID 16959962
- ↑ Yang Y, et al. (2010) Identifying cooperative transcription factors by combining ChIP-chip data and knockout data. Cell Res 20(11):1276-8 SGD PMID 20975739
See Help:Categories on how to add the wiki page for this gene to a Category </protect>