Difference between revisions of "SGD Newsletter, December 2025"
(→Final Release of WormBase) |
(→Final Release of WormBase) |
||
| Line 69: | Line 69: | ||
WormBase announced their final release, WS298, on November 27, 2025. The existing [https://wormbase.org WormBase] website will continue provide access to this archived WS298 release of WormBase, and the content hosted there will no longer be updated except for essential maintenance to ensure continued availability. Ongoing curation, annotation, data updates, and new data for ''Caenorhabditis elegans'' will now be managed through the Alliance of Genome Resources. Users should refer to the Alliance for the most current worm datasets and continued resource development. Read more on the [https://blog.wormbase.org/2025/07/23/announcing-the-final-release-of-wormbase/ WormBase blog]. | WormBase announced their final release, WS298, on November 27, 2025. The existing [https://wormbase.org WormBase] website will continue provide access to this archived WS298 release of WormBase, and the content hosted there will no longer be updated except for essential maintenance to ensure continued availability. Ongoing curation, annotation, data updates, and new data for ''Caenorhabditis elegans'' will now be managed through the Alliance of Genome Resources. Users should refer to the Alliance for the most current worm datasets and continued resource development. Read more on the [https://blog.wormbase.org/2025/07/23/announcing-the-final-release-of-wormbase/ WormBase blog]. | ||
| + | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 11:29, 11 December 2025
About this newsletter:
This is the December 2025 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community.
Contents
Support SGD this Holiday Season
Budget cuts from NIH continue to strain SGD’s finances. Despite our efforts at reducing costs, we still have significant ongoing budgetary challenges. Donations are now critical for our work to continue. Your generous gift to SGD enables us to continue providing essential information for your research and teaching efforts. We are now able to accept gifts via credit card. To contribute using a credit card, please use this form: give.stanford.edu.
- Under ‘Direct your gift,’ select ‘Other Stanford Designation’ from the pulldown menu
- In the ‘Other’ text box, specify SGD by including the text “Saccharomyces Genome Database - Account : GHJKO, Genetics : WAZC”
- Complete the form and payment to donate to SGD
If you’d like to contribute by check, please contact us at: sgd-helpdesk@lists.stanford.edu
We thank you in advance for your support!
New at SGD: Links to a Comprehensive Overview of Yeast Libraries
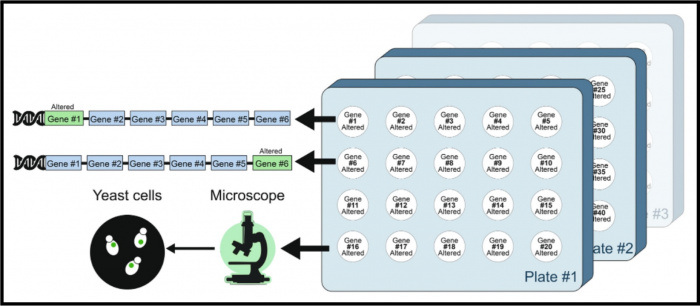
Baruch et al. have compiled and published A comprehensive overview of yeast libraries and their role in advancing cell biology, which describes a comprehensive list of genome-wide collections of Saccharomyces cerevisiae. They have grouped similar strain libraries together and have curated detailed descriptions, mating types, genotypes, references, and genome coverage. This information can help researchers identify strain libraries of interest to them. All of this information can also be found in tables on LibrarYeast. We have added links to these tables under the Resources section of individual Protein, Phenotype, and Interaction tabs.
Retirement of Mike Cherry
This year, we celebrate and thank J. Michael “Mike” Cherry as he retires after decades of visionary leadership of the Saccharomyces Genome Database. From the earliest days of the yeast genome to today’s multi‑omic era, Mike helped shape SGD into the trusted, rigorously curated resource at the heart of the yeast community. His commitment to high‑quality data, interoperability, and open science empowered discoveries around the world, and his mentorship nurtured generations of curators, developers, and collaborators. Mike’s steady guidance, big‑picture thinking, and relentless focus on serving researchers have left a lasting legacy that will continue to guide SGD’s mission.
On November 14th, many past and current Cherry lab members gathered at Stanford to celebrate Mike’s career. Thank you to everyone who attended, and especially to everyone that shared stories or memories from the past 3 decades of SGD. Gavin Sherlock is SGD's new PI, and he is hardly a stranger to SGD! Mike and Gavin published their first article together about SGD in Science all the way back in 1998: Chervitz et al., Comparison of the Complete Protein Sets of Worm and Yeast: Orthology and Divergence.
On behalf of the SGD team and the global yeast community, we extend our deepest gratitude to Mike and wish him joy and fulfillment in the next chapter.
Thank you, Mike, for everything.
Reminder: Submit Data Form
If you're itching for something to do over the winter break, have you checked your newly published data at SGD lately? SGD can use your help! Authors can submit data and information about their publications by pointing us to novel results, datasets (we appreciate GEO accession IDs!), or other important information, using SGD's simple “Submit Data" form (https://www.yeastgenome.org/submitData).
Alliance of Genome Resources News
Alliance of Genome Resources – Latest Release 8.2.0
The 8.2.0 release includes data refreshes from each of the model organism source databases as well as various backend improvements. The sequence viewers on the gene and allele pages have been restored on gene pages, in the Alleles and Variants and Sequence Feature Viewer sections, and on allele pages, in the Genomic Variant Information section (see for example the mouse gene Pten and mouse allele for Pten). Also, on the gene page Alleles and Variants section, interactions between the variant sequence viewer and the alleles/variants table have been restored. Website infrastructure is upgraded to React 19 and migrated to Vite.
Alliance Webinars: MicroPublication Biology
View the video of the latest Alliance of Genome Resources webinar: Introduction to MicroPublication Biology On November 20, Tim Schedl, microPublication Biology Editorial Board member, presented an overview of the MicroPublication journal which publishes brief scholarly reports of research findings based on data presented in a single figure. Webinars are also planned for 2026, in January, February, and March. See the Event Calendar for the schedule of upcoming Alliance office hours and webinars.
FlyBase End of Funding
The termination of the NIH/NHGRI FlyBase grant has placed the long-term sustainability of FlyBase at risk. However, thanks to the generous support of several key individuals and institutions, FlyBase has announced that they will remain operational through the coming year. Looking ahead, ensuring FlyBase’s sustainability beyond the next year – and successfully integrating with the Alliance – will require new funding sources. FlyBase, like SGD, is asking for continued donations from any and all Fly supporters. For more information on how to support FlyBase, see the FlyBase wiki:
- European labs: Please consider contributing to the Cambridge, U.K. FlyBase group
- U.S. and other non-European labs: Please consider contributing to the U.S. FlyBase groups
Final Release of WormBase
WormBase announced their final release, WS298, on November 27, 2025. The existing WormBase website will continue provide access to this archived WS298 release of WormBase, and the content hosted there will no longer be updated except for essential maintenance to ensure continued availability. Ongoing curation, annotation, data updates, and new data for Caenorhabditis elegans will now be managed through the Alliance of Genome Resources. Users should refer to the Alliance for the most current worm datasets and continued resource development. Read more on the WormBase blog.
microPublications – Latest Yeast Papers
microPublication Biology is part of the emerging genre of rapidly-published research communications. microPublications publishes brief, novel findings, negative and/or reproduced results, and results which may initially lack a broader scientific narrative. Each article is peer-reviewed, assigned a DOI, and indexed through PubMed and PubMedCentral. Consider microPubublications when you have a result that doesn’t necessarily fit into a larger story, but will be of value to others. Latest yeast microPublications:
- King A, Emery TM, Reese K, Tinsley HN (2025) Yeast gene of unknown function YGL081W is involved in DNA damage response. MicroPubl Biol 2025
- Avogo EW, Burlingame NA, Badenahalli Narasimhaiah S, Delorme-Axford E (2025) Bioinformatics analysis identifies Mot2 protein as a potential regulator of autophagy in Saccharomyces cerevisiae. MicroPubl Biol 2025
- Nnabuenyi N, Sands MA, Camlin NJ (2025) Specific auxin and medium combinations alter Saccharomyces cerevisiae growth. MicroPubl Biol 2025
- Sasahara M, Matsumoto S, Tamura Y (2025) Dnm1 Is Required for the Focal Clustering of Fis1 on the Mitochondrial Outer Membrane. MicroPubl Biol 2025
- Dutca LM, Freed EF, Baserga SJ (2025) Yeast pre-rRNA is processed at the A' site. MicroPubl Biol 2025
- Schiemann AH, Sarwar M, Sattlegger E (2025) Functional Analysis of the Yeast Counterpart to the Human GCN2 p.Glu738_Asp739insArgArg Variant. MicroPubl Biol 2025
- Torvi JR, Wong J, Drubin DG, Barnes G (2025) Stu2 is required for plus-end directed chromosome transport along microtubules during metaphase in Saccharomyces cerevisiae. MicroPubl Biol 2025
- VanderVen K, Butcher C, Fokine R, Li J (2025) Pep12 is important for proteasome microautophagy under low glucose conditions. MicroPubl Biol 2025
- Tang L, AlKaabi A, Meyer D (2025) Copper Homeostasis is influenced by Ics3 in Saccharomyces cerevisiae. MicroPubl Biol 2025
- Taguchi S, Matsuzawa R, Suda Y, Irie K, Ozaki H (2025) Investigating the effects of liquid handling robot pipetting speed on yeast growth and gene expression using growth assays and RNA-seq. MicroPubl Biol 2025
- Takesue H, Okada S, Ito T (2025) Long-read plasmid sequencing strategy for evaluating intrinsic instability of tandem gene arrays. MicroPubl Biol 2025
Upcoming Conferences & Courses
- 33rd Fungal Genetics Conference
- March 17 to March 22, 2026 -
- Asilomar Conference Grounds, Pacific Grove, CA
- Yeast Genetics Meeting 2026
- June 13 to June 17, 2026 -
- Asilomar Conference Grounds, Pacific Grove, CA
- 39th International Specialized Symposium on Yeasts (ISSY39)
- Nov 8 - 12, 2026
- Jeongdong 1928 Art Center, Seoul, KR
Happy Holidays from SGD!
We want to take this opportunity to wish you and your family, friends, and lab mates the best during the upcoming holidays. Stanford University will be closed for two weeks starting end of day Friday, December 19th, and reopening on Monday, January 5th, 2026. Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will resume responding to user requests and questions in the new year. Note: If you wish to receive this newsletter via email, please contact the SGD Help Desk at sgd-helpdesk@lists.stanford.edu.