Difference between revisions of "YCR095C"
(→Ethanol) |
(→Ethanol) |
||
| Line 32: | Line 32: | ||
[[Image:Best Ethanol KO graph.png|Graph|center|700px|YJL133C-A]] | [[Image:Best Ethanol KO graph.png|Graph|center|700px|YJL133C-A]] | ||
| − | + | ||
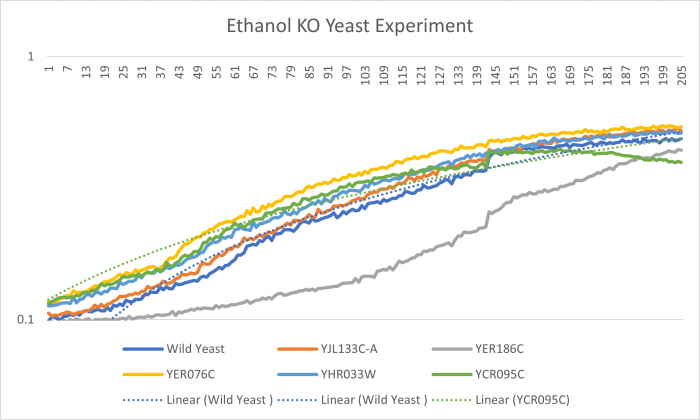
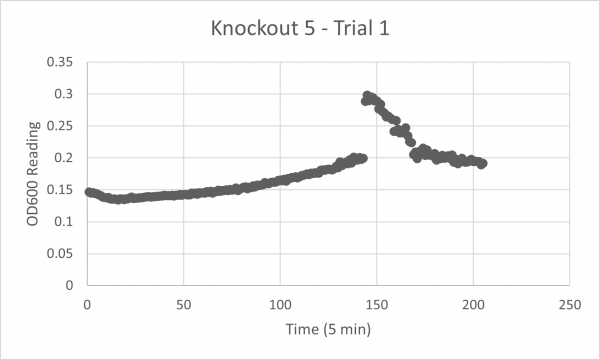
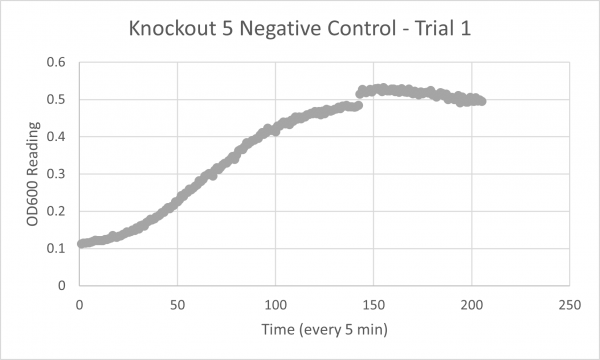
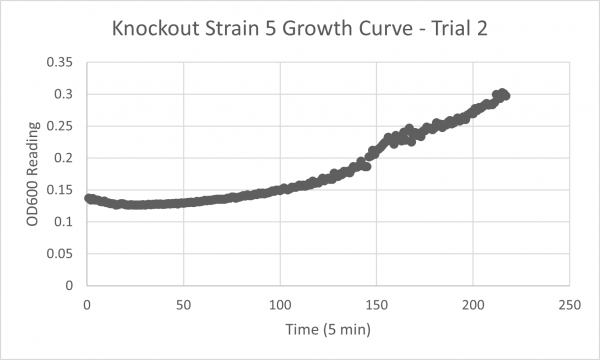
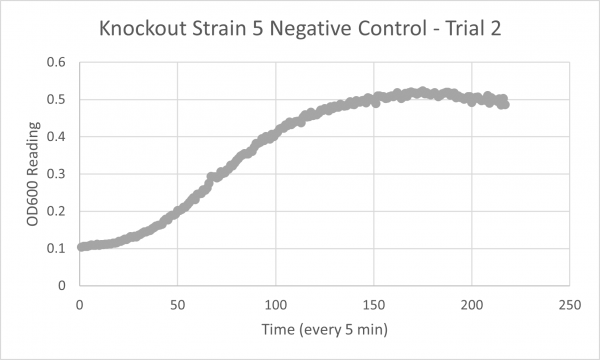
| + | YJL133C-A's line lies in the middle of the overall growth curve. The doubling time is 67.78 | ||
===[[UW-Stout/Caffeine_FA22|Caffeine]]=== | ===[[UW-Stout/Caffeine_FA22|Caffeine]]=== | ||
Revision as of 08:29, 17 December 2022
Share your knowledge...Edit this entry! <protect>
| Systematic name | YCR095C |
| Gene name | OCA4 |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr III:289258..288170 |
| Primary SGDID | S000000691 |
Description of YCR095C: Cytoplasmic protein required for replication of Brome mosaic virus in S. cerevisiae, which is a model system for studying replication of positive-strand RNA viruses in their natural hosts[1][2]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
Ethanol
YJL133C-A's line lies in the middle of the overall growth curve. The doubling time is 67.78
Caffeine
Interpretation
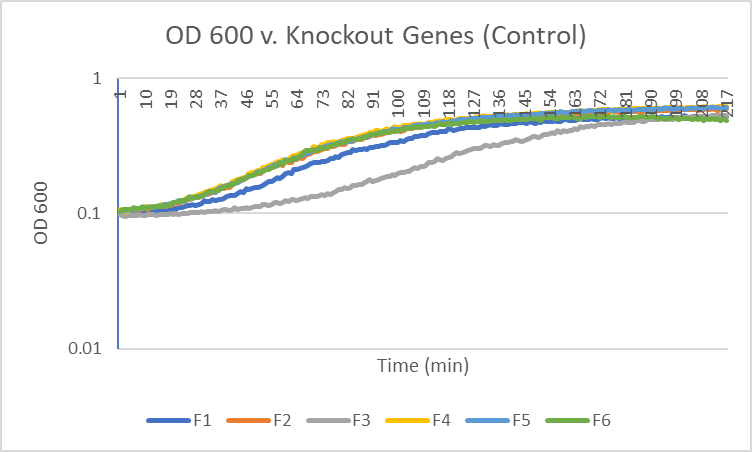
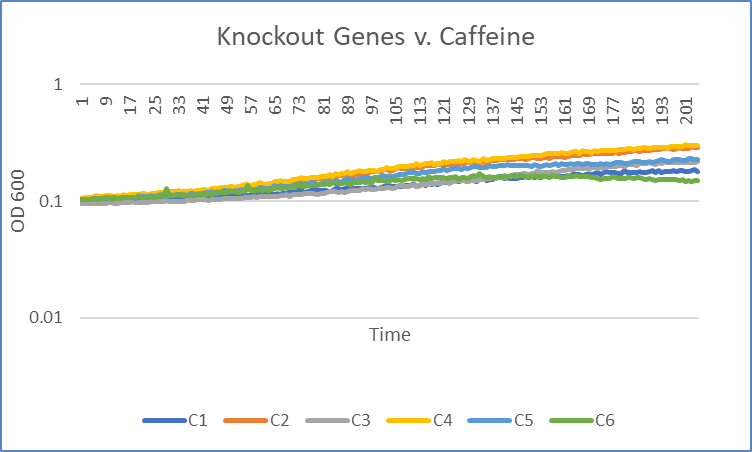
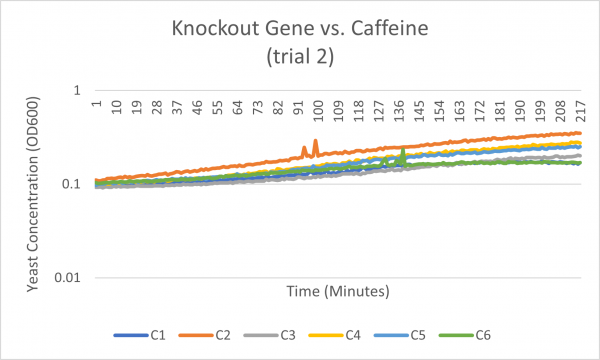
The average doubling time for the YCR095C (C6) gene in standard conditions was 99 minutes. Under caffeine stress, the average doubling time was 344 minutes. That is a 237% increase in doubling time which means that cell growth was negatively affected (slowed down) by this type of stress.
<protect>
References
See Help:References on how to add references
- ↑ Huh WK, et al. (2003) Global analysis of protein localization in budding yeast. Nature 425(6959):686-91 SGD PMID 14562095
- ↑ Kushner DB, et al. (2003) Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A 100(26):15764-9 SGD PMID 14671320
See Help:Categories on how to add the wiki page for this gene to a Category </protect>
UW Stout/FA 2022 Glycerol Media
Introduction
Following the UW-Stout/Glycerol FA22 protocol, 5 knockout strains of yeast cell were tortured within a glycerol media solution.
Results
The following doubling times were calculated:
- Glycerol Trial 1: NA
- Negative Control Trial 1: 202 min
- Glycerol Trial 2: 689 min
- Negative Control Trial 2: 176 min
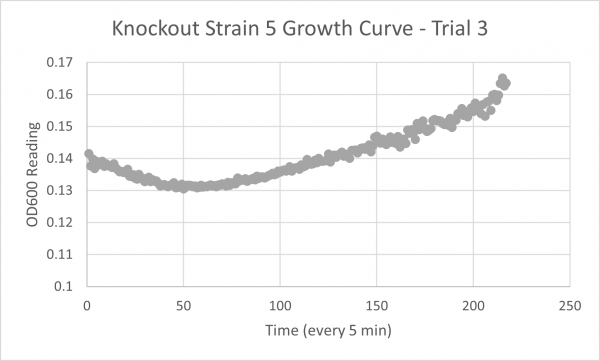
- Glycerol Trial 3: 2868 min
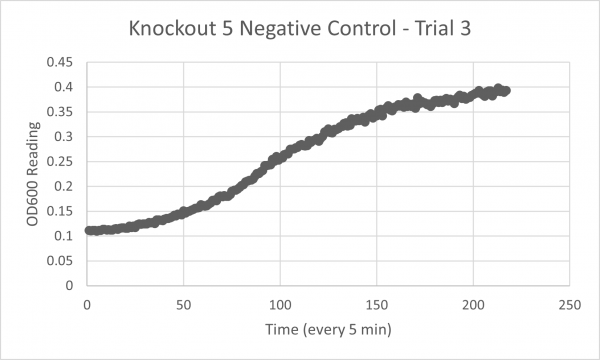
- Negative Control Trial 3: 264 min
Analysis/Conclusion
In the strains of yeast where the YCR095C gene was knocked out, a significant increase in doubling time can be seen. Disregarding trial 1 due to an unexpected collection error, the doubling times in the 2nd and 3rd trials, where the sample was kept in a 3% gliserol growth medium, showed significant results. In the second trial, the negative control had a doubling time of 176 while the glycerol sample had a doubling time of 689 min; this is an increase that is comparable to the pilot, where the doubling time doubled from the negative control to the glycerol sample. What we can conclude from this is that the samples in the glycerol media are only able to divide at a rate of about ½ to ⅓ of the rate of the glucose samples. Trial 3 shows a very exaggerated form of the data seen in Trial 2. This can be written up to error in preparing the samples, but the fact that the negative control for Trial 3 has a doubling time of 264 min while the one for the glycerol sample is 2868 min still shows that the cells have a much harder time dividing in the glycerol solution.