Difference between revisions of "UW-Stout/Formamide SP22"
(→Procedure) |
(→Procedure) |
||
| Line 43: | Line 43: | ||
## Shake before read: 30 seconds | ## Shake before read: 30 seconds | ||
# Transfer the assay plate to the reader and read for 24 hours | # Transfer the assay plate to the reader and read for 24 hours | ||
| + | # Record data | ||
===Results=== | ===Results=== | ||
Revision as of 18:44, 9 May 2022
Contents
Wild Yeast Cell Pilot Procedure
Caution
Neat Formamide is harmful to the eyes, if swallowed, inhaled, or absorbed through the skin.
Materials / Equipment
- Neat Formamide, 33ul
- Sterile water, 567ul
- 0.2ml flat cap PCR tube, 13
- Well plate
- Wild yeast cells, 600ul
- Molecular Devices SpectraMax Plus 384 Microplate Reader
- Micropipet 100ul
- Micropipet tips
Formamide Concentrations
- 0.5ul formamide + 49.5ul sterile water
- 1.0ul formamide + 49.0ul sterile water
- 1.5ul formamide + 48.5ul sterile water
- 2.0ul formamide + 48.0ul sterile water
- 2.5ul formamide + 47.5ul sterile water
- 3.0ul formamide + 47.0ul sterile water
- 3.5ul formamide + 46.5ul sterile water
- 4.0ul formamide + 46.0ul sterile water
- 4.5ul formamide + 45.5ul sterile water
- 5.0ul formamide + 45.0ul sterile water
- 5.5ul formamide + 44.5ul sterile water
- 0.0ul formamide + 50.0ul sterile water
Procedure
- Obtain the neat formamide (refrigerated) and sterile water
- Obtain and label 12 PCR tubes 1-12
- Obtain one more PCR tube to transfer 33ul of neat formamide inside, can put excess in
- Using the 13th PCR tube, pipet appropriate amount of formamide and sterile water into the labeled PCR tubes according to number
- In a sterile environment, pipet the liquid (50ul) from the PCR tube into the well cell
- Vortex the yeast culture briefly to resuspend the yeast cells, and then pipet 50ul of the wild yeast cells into each well in addition to the formamide
- Set up the plate reader as follows:
- Temperature: 30 degrees Celsius
- Mode: kinetic
- Wavelength: 600 nm
- Interval: 5 minutes
- Total run time: 24 hours
- Shake before read: 30 seconds
- Transfer the assay plate to the reader and read for 24 hours
- Record data
Results
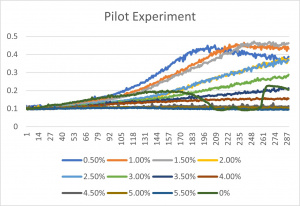
- The x-axis corresponds to the concentrations listed up above (ex: B1 corresponds to the 0.5%)
- 0% is an experimental error- since it has a concentration of 0% formamide anyway, it was left in the final graph
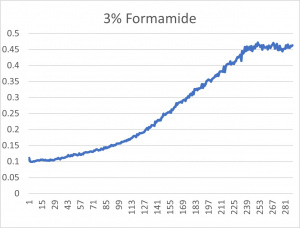
- The 3% formamide used in the pilot experiment and represented as B-5 (in the top-most picture) was chosen to be used on the transformed yeast cells
- This was deduced due to the position of B-5
- B-5 was able to grow, but didn't grow too much- simply put, it was in the middle
Knockout Yeast Cell Gene
Materials / Equipment
- Neat formamide, 18ul
- Sterile water, 423ul
- 0.2ml flat cap PCR tube 10
- Well plate
- Knock out yeast cells
- Molecular Devices SpectraMax Plus 384 Microplate Reader
- Micropipet 100ul
- Micropipet tips
Procedure
- Obtain neat formamide (refrigerated) and sterile water
- Obtain and label PCR tubes 1-9
- Pipet approximately 18ul formamide into a 10th PCR tube, can put excess formamide in
- Using the 10th PCR tube, pipet 3.0ul of formamide into each PCR tube (labeled 1-9)
- Pipet 47.0ul of sterile water into each of the numbered PCR tubes
- In a sterile environment, pipet 50ul of liquid from the PCR tubes into well cells. Vortex each yeast culture strain briefly to resuspend the cells, and then pipet 50ul of each knockout strain into the appropriate well cells
- Set up the plate reader as follows:
- Temperature: 30 degrees Celsius
- Mode: kinetic
- Wavelength: 600 nm
- Interval: 5 minutes
- Total run time: 24 hours
- Shake before read: 30 seconds
- Transfer the assay plate to the reader and read for 24 hours
- Record data
Results
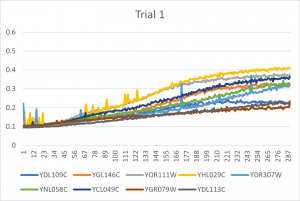
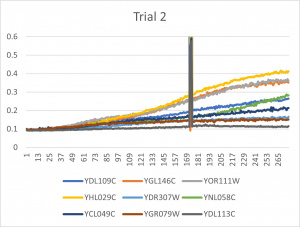
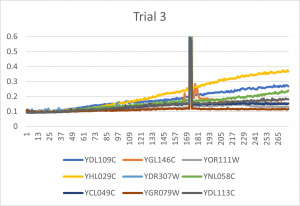
- Ran three trials, trial one through three is posted up above in picture form
- The dark line in the middle of the graph is a scanning error from the Molecular Devices SpectraMax Plus 384 Microplate Reader
- Computed doubling times from the average times of the three trials for further analyzation