Difference between revisions of "YER186C"
SGDwikiBot (talk | contribs) (Automated import of articles) |
|||
| (41 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- PLEASE DO NOT EDIT HERE. USE THE SECTION EDIT LINKS ON THE RIGHT MARGIN--> | <!-- PLEASE DO NOT EDIT HERE. USE THE SECTION EDIT LINKS ON THE RIGHT MARGIN--> | ||
{{PageTop}} | {{PageTop}} | ||
| + | <protect> | ||
{|{{Prettytable}} align = 'right' width = '200px' | {|{{Prettytable}} align = 'right' width = '200px' | ||
|- | |- | ||
| − | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http:// | + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http://www.yeastgenome.org/cgi-bin/locus.pl?dbid=S000000988 YER186C] |
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | ||
| Line 12: | Line 13: | ||
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | ||
| − | |nowrap| Chr V: | + | |nowrap| Chr V:562625..561705 |
|- | |- | ||
| − | | | + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Primary SGDID''' || S000000988 |
|} | |} | ||
<br> | <br> | ||
| − | '''Description of | + | '''Description of YER186C:''' Putative protein of unknown function<ref name='S000114148'>SGD (2006) No information available |
| + | {{SGDpaper|S000114148}} PMID </ref> | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
| − | + | <br> | |
| + | </protect> | ||
__TOC__ | __TOC__ | ||
==Community Commentary== | ==Community Commentary== | ||
{{CommentaryHelp}} | {{CommentaryHelp}} | ||
| + | === DNA and RNA Details === | ||
| + | [[Category:Topic:DNA and RNA Details]] | ||
| + | ==== Other DNA and RNA Details ==== | ||
| + | [[Category:Topic:DNA and RNA Details:Other DNA and RNA Details]] | ||
| + | '''Other Topic''': expression [[Category:Topic:expression]] | ||
| + | |||
| + | Specifically higher expression in phosphorus limited chemostat cultures versus phosphorus excess. <ref name='S000073646'>Boer VM, et al. (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278(5):3265-74 {{SGDpaper|S000073646}} PMID 12414795</ref> <ref name = 'CAset9153-2003-07-25'>submitted by [http://db.yeastgenome.org/cgi-bin/colleague/colleagueSearch?id=9153 Viktor Boer] on 2003-07-25</ref> | ||
| + | |||
| + | {{ShortCenteredHR}} | ||
| + | |||
| + | ===[[UW-Stout/Caffeine_FA22|Caffeine]]=== | ||
| + | |||
| + | |||
| + | [[Image:F_graph.png|100pxs|Control]] | ||
| + | |||
| + | [[Image:Knockout_v._caffeine_trial1_Viv.png|100pxs|Trial 1]] | ||
| + | |||
| + | [[Image:Knockout_gene_v._Caffeine_trial_2.png|600px|Trial 2]] | ||
| + | |||
| + | ====Interpretation==== | ||
| + | |||
| + | The average doubling time for the YEr186C (C3) gene in standard conditions was 87 minutes. Under caffeine stress, the average doubling time was 181 minutes. That is a 108% increase in doubling time which means that cell growth was negatively affected (slowed down) by this type of stress. | ||
| + | |||
| + | ===[[UW-Stout/Ethanol_FA22|Ethanol]]=== | ||
| + | [[Image:Best Ethanol KO graph.png|Graph|center|700px|YJL133C-A]] | ||
| + | YER186C is our outlier for this Ethanol experiment. The Ethanol dramatically decreased cell growth. The doubling time is 189.60 | ||
| + | |||
| + | ===Hydrogen Peroxide=== | ||
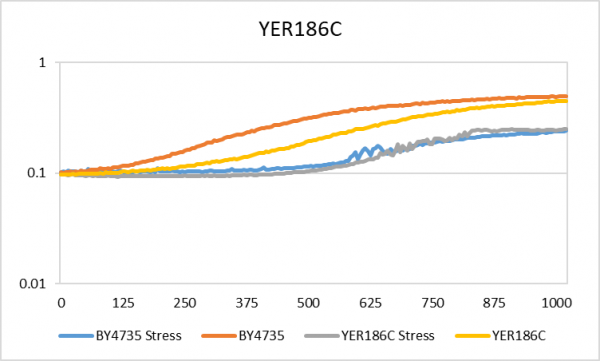
| + | [[Image:YER186C.png|center|thumb|600px|The x-axis is time in minutes and the y-axis is OD600]] | ||
| + | This is the link to the protocol page: [[UW-Stout/Hydrogen Peroxide FA22]]. | ||
| + | |||
| + | The doubling time for the YER186C gene under stressed conditions is 539 minutes and without stress is 314 minutes. The wild type yeast under stress had a 598 minute doubling time and with no stress there was a 229 minute doubling time. The doubling time for both yeast strains with and without stress was very similar, so I do not not expect YER186C to play a large role in yeast growth when hydrogen peroxide is present. | ||
| + | |||
| + | |||
| + | <!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | ||
| + | <!-- | ||
| + | Specifically higher expression in carbon limited chemostat cultures versus carbon excess. | ||
| + | <ref>Boer VM, et al. (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. | ||
| + | J Biol Chem 278(5):3265-74</ref> | ||
| + | --> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <protect> | ||
==References== | ==References== | ||
<!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | <!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | ||
{{RefHelp}} | {{RefHelp}} | ||
| + | </protect> | ||
| + | |||
| + | ==UW Stout/FA 2022 Glycerol Media== | ||
| + | ===Introduction=== | ||
| + | Following the [[UW-Stout/Glycerol FA22]] protocol, 5 knockout strains of yeast cell were tortured within a glycerol media solution. | ||
| + | ===Results=== | ||
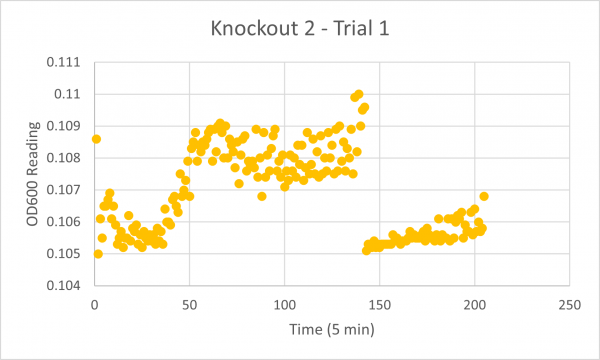
| + | [[Image:Knockout_2_Trial_1.png|600px]] | ||
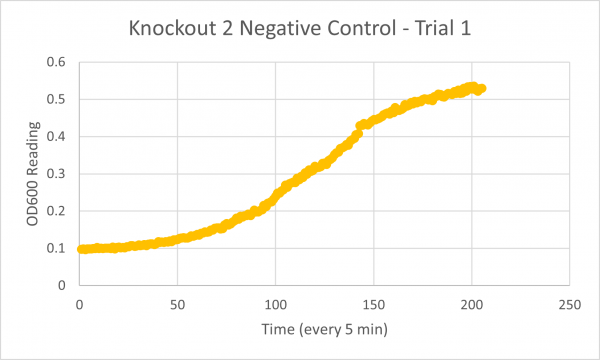
| + | [[Image:Negative_Control_KO2_Trial_1.png|600px]] | ||
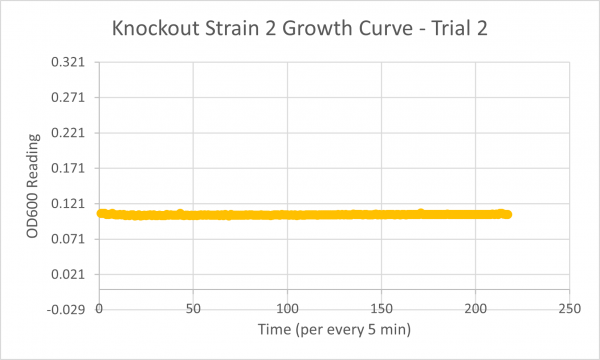
| + | [[Image:Knockout_2_Trial_2.png|600px]] | ||
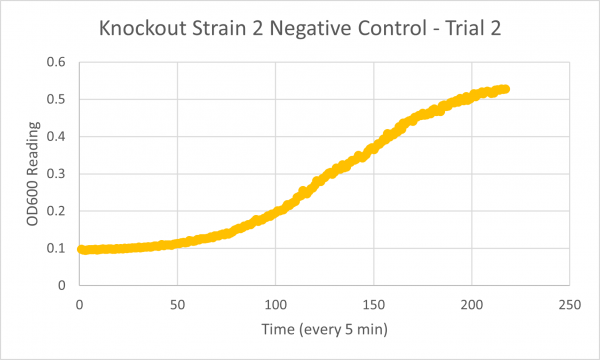
| + | [[Image:Negative_Control_KO2_T2.png|600px]] | ||
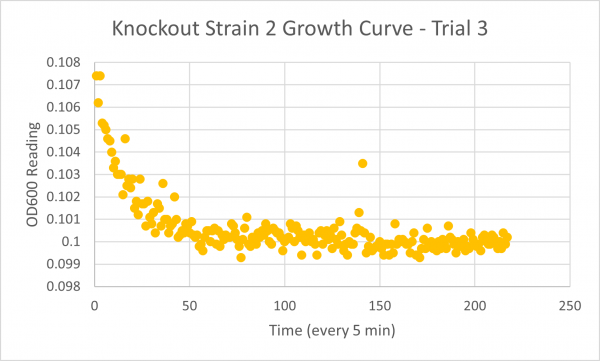
| + | [[Image:Growth_Curve_KO2_Trial_3.png|600px]] | ||
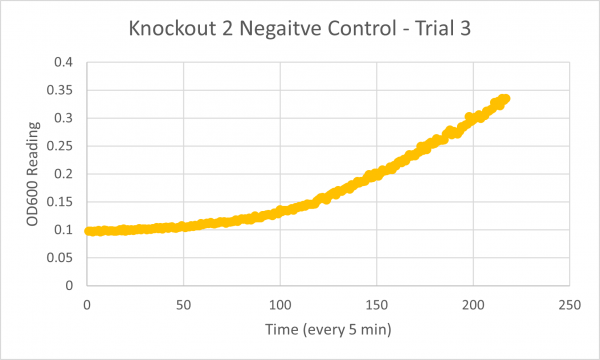
| + | [[Image:Negative_Control_KO2_Trial_3.png|600px]] | ||
| + | |||
| + | The following doubling times were calculated: | ||
| + | * Glycerol Trial 1: NA | ||
| + | * Negative Control Trial 1: 173min | ||
| + | * Glycerol Trial 2: NA | ||
| + | * Negative Control Trial 2: 287min | ||
| + | * Glycerol Trial 3: NA | ||
| + | * Negative Control Trial 3: 262min | ||
| + | |||
| + | ===Analysis/Conclusion=== | ||
| + | The results from glycerol torture on the YER186C knockout were quite interesting. All 3 graphs looked ridiculous, but there is one point that is clear: the yeast cells did not grow very successfully in the glycerol media without this gene. Trial 1's data was all around a little scattered and the jump near the end was a bit concerning considering it happened in every single experiment- so we weren't very confident in the results. However, in Trial 2 it continued to be weird... The OD600 reading did not seem to go up at all! This could mean the cells are all dead or not reproducing. This is intriguing data! The negative control graph for this trial even looked normal which leads us to believe that the flatlined data wasn't just a slip up. Trial 3, like trial 1, also seemed like a bust. It was interesting that the data showed a significant decrease in the original cells, but one thing is for certain: the yeast was definitely not reproducing and was most likely dead. | ||
| + | |||
| + | ==UW Stout/FA 2022 Bud Scars== | ||
| + | Cells that had the gene YER186C had an average of 2.82 scars per cell. Out of 100 cells counted, 282 bud scars were counted. The wild yeast BY4735 had an average of 2.46 bud scars per cell (100 cells counted, 246 bud scars). This indicates that the knocked-out gene slightly sped up the reproduction rate of these cells. | ||
| + | [[File:Yeast_2(2)_7_8.png]] | ||
| + | ''picture does not include all cells that were counted and added for a visual of bud scars'' | ||
Latest revision as of 17:40, 20 December 2022
Share your knowledge...Edit this entry! <protect>
| Systematic name | YER186C |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr V:562625..561705 |
| Primary SGDID | S000000988 |
Description of YER186C: Putative protein of unknown function[1]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
DNA and RNA Details
Other DNA and RNA Details
Other Topic: expression
Specifically higher expression in phosphorus limited chemostat cultures versus phosphorus excess. [2] [3]
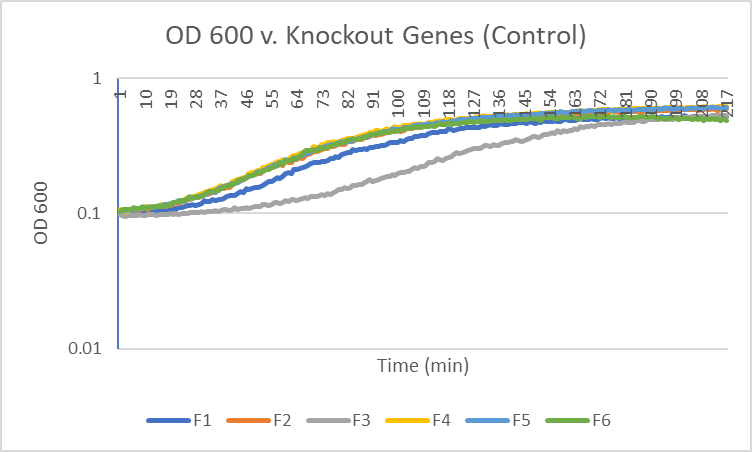
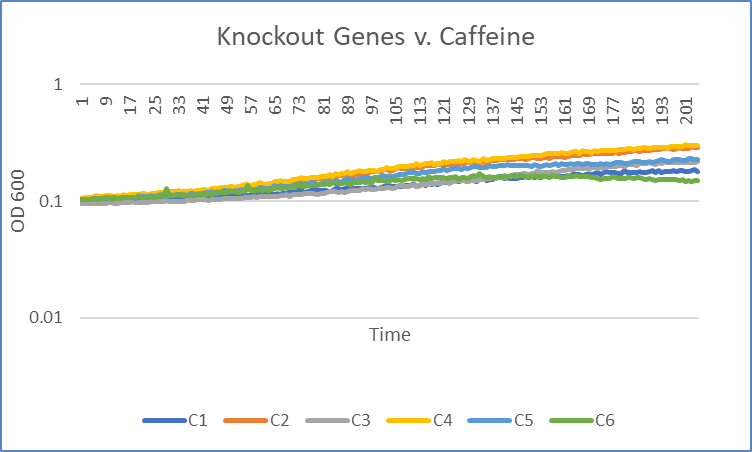
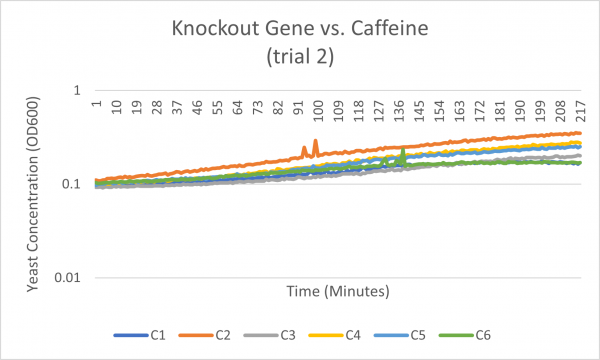
Caffeine
Interpretation
The average doubling time for the YEr186C (C3) gene in standard conditions was 87 minutes. Under caffeine stress, the average doubling time was 181 minutes. That is a 108% increase in doubling time which means that cell growth was negatively affected (slowed down) by this type of stress.
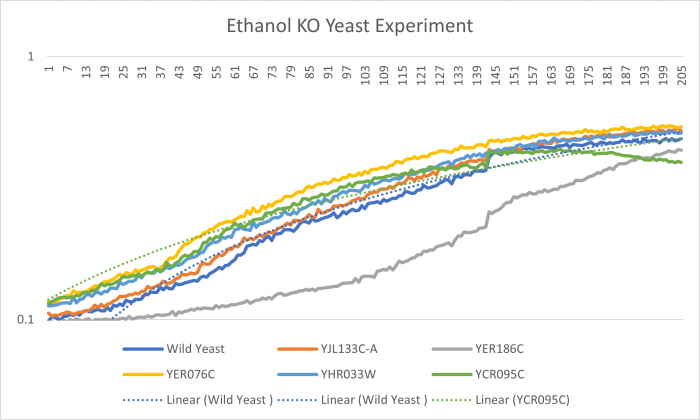
Ethanol
YER186C is our outlier for this Ethanol experiment. The Ethanol dramatically decreased cell growth. The doubling time is 189.60
Hydrogen Peroxide
This is the link to the protocol page: UW-Stout/Hydrogen Peroxide FA22.
The doubling time for the YER186C gene under stressed conditions is 539 minutes and without stress is 314 minutes. The wild type yeast under stress had a 598 minute doubling time and with no stress there was a 229 minute doubling time. The doubling time for both yeast strains with and without stress was very similar, so I do not not expect YER186C to play a large role in yeast growth when hydrogen peroxide is present.
<protect>
References
See Help:References on how to add references
- ↑ SGD (2006) No information available SGD PMID
- ↑ Boer VM, et al. (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278(5):3265-74 SGD PMID 12414795
- ↑ submitted by Viktor Boer on 2003-07-25
See Help:Categories on how to add the wiki page for this gene to a Category </protect>
UW Stout/FA 2022 Glycerol Media
Introduction
Following the UW-Stout/Glycerol FA22 protocol, 5 knockout strains of yeast cell were tortured within a glycerol media solution.
Results
The following doubling times were calculated:
- Glycerol Trial 1: NA
- Negative Control Trial 1: 173min
- Glycerol Trial 2: NA
- Negative Control Trial 2: 287min
- Glycerol Trial 3: NA
- Negative Control Trial 3: 262min
Analysis/Conclusion
The results from glycerol torture on the YER186C knockout were quite interesting. All 3 graphs looked ridiculous, but there is one point that is clear: the yeast cells did not grow very successfully in the glycerol media without this gene. Trial 1's data was all around a little scattered and the jump near the end was a bit concerning considering it happened in every single experiment- so we weren't very confident in the results. However, in Trial 2 it continued to be weird... The OD600 reading did not seem to go up at all! This could mean the cells are all dead or not reproducing. This is intriguing data! The negative control graph for this trial even looked normal which leads us to believe that the flatlined data wasn't just a slip up. Trial 3, like trial 1, also seemed like a bust. It was interesting that the data showed a significant decrease in the original cells, but one thing is for certain: the yeast was definitely not reproducing and was most likely dead.
UW Stout/FA 2022 Bud Scars
Cells that had the gene YER186C had an average of 2.82 scars per cell. Out of 100 cells counted, 282 bud scars were counted. The wild yeast BY4735 had an average of 2.46 bud scars per cell (100 cells counted, 246 bud scars). This indicates that the knocked-out gene slightly sped up the reproduction rate of these cells.
 picture does not include all cells that were counted and added for a visual of bud scars
picture does not include all cells that were counted and added for a visual of bud scars