Difference between revisions of "SGD Newsletter, December 2024"
(→Give a Gift / Support SGD) |
(→SGD’s new GENETICS publication) |
||
| (16 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Category: Newsletter]] | [[Category: Newsletter]] | ||
'''About this newsletter:'''<br> | '''About this newsletter:'''<br> | ||
| − | This is the | + | This is the December 2024 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features from SGD and to foster communication within the yeast community. |
==AlphaFold protein structures now on SGD protein pages== | ==AlphaFold protein structures now on SGD protein pages== | ||
| Line 7: | Line 7: | ||
We are thrilled to announce that we have now integrated [https://alphafold.ebi.ac.uk AlphaFold] protein structures into our [https://www.yeastgenome.org/locus/set1/protein protein pages]! This cutting-edge addition provides detailed, high-accuracy 3D models of protein structures, offering invaluable insights into protein function and interactions. Researchers can now explore these comprehensive structural predictions directly within SGD, facilitating advanced studies in molecular biology and bioinformatics. Dive into the new AlphaFold protein structures and elevate your research with this powerful tool! | We are thrilled to announce that we have now integrated [https://alphafold.ebi.ac.uk AlphaFold] protein structures into our [https://www.yeastgenome.org/locus/set1/protein protein pages]! This cutting-edge addition provides detailed, high-accuracy 3D models of protein structures, offering invaluable insights into protein function and interactions. Researchers can now explore these comprehensive structural predictions directly within SGD, facilitating advanced studies in molecular biology and bioinformatics. Dive into the new AlphaFold protein structures and elevate your research with this powerful tool! | ||
| − | AlphaFold, developed by [https://deepmind.google/technologies/alphafold/ DeepMind], is an AI program that accurately predicts protein structures from amino acid sequences, enabling visualization of protein conformations. The predicted structures can be accessed through the [https://www.rcsb.org/ Protein Data Bank (PDB)] and [https://www.rcsb.org/ AlphaFold Protein Structure Database]. | + | AlphaFold, developed by [https://deepmind.google/technologies/alphafold/ DeepMind], is an AI program that accurately predicts protein structures from amino acid sequences, enabling visualization of protein conformations. |
| + | |||
| + | The predicted structures can be accessed through the [https://www.rcsb.org/ Protein Data Bank (PDB)] and [https://www.rcsb.org/ AlphaFold Protein Structure Database]. | ||
Thanks to Kim Rutherford and Val Wood of [https://www.pombase.org Pombase] for tips about adding AlphaFold structures to SGD. | Thanks to Kim Rutherford and Val Wood of [https://www.pombase.org Pombase] for tips about adding AlphaFold structures to SGD. | ||
| − | |||
| − | |||
==YeastMine data available in AllianceMine== | ==YeastMine data available in AllianceMine== | ||
| Line 21: | Line 21: | ||
In July 2024, YeastMine was discontinued due to ongoing cuts in funding at SGD. However, we have moved the YeastMine data into [https://www.alliancegenome.org/bluegenes/alliancemine AllianceMine], hosted by the [https://www.alliancegenome.org/ Alliance of Genome Resources], of which SGD is a founding member. You can even access lists like [https://www.alliancegenome.org/bluegenes/alliancemine/results/ALL_Verified_Uncharacterized_Dubious_ORFs “ALL_Verified_Uncharacterized_Dubious_ORFs”] through the [https://www.alliancegenome.org/bluegenes/alliancemine/lists AllianceMine ''Lists''] just as you could in the previous versions of YeastMine. Ensure any bookmarks to YeastMine have been updated to match the new URL: '''https://www.alliancegenome.org/bluegenes/alliancemine'''. User documentation for the new YeastMine interface is [http://intermine.org/intermine-user-docs/ available from InterMine]. | In July 2024, YeastMine was discontinued due to ongoing cuts in funding at SGD. However, we have moved the YeastMine data into [https://www.alliancegenome.org/bluegenes/alliancemine AllianceMine], hosted by the [https://www.alliancegenome.org/ Alliance of Genome Resources], of which SGD is a founding member. You can even access lists like [https://www.alliancegenome.org/bluegenes/alliancemine/results/ALL_Verified_Uncharacterized_Dubious_ORFs “ALL_Verified_Uncharacterized_Dubious_ORFs”] through the [https://www.alliancegenome.org/bluegenes/alliancemine/lists AllianceMine ''Lists''] just as you could in the previous versions of YeastMine. Ensure any bookmarks to YeastMine have been updated to match the new URL: '''https://www.alliancegenome.org/bluegenes/alliancemine'''. User documentation for the new YeastMine interface is [http://intermine.org/intermine-user-docs/ available from InterMine]. | ||
| − | == | + | ==Help maintain your genomic databases! Sign open letter supporting funding for biodata resources== |
[[Image:FkAzFtQWIAExhDl.jpeg|link=https://globalbiodata.org/open-letter-campaign/|thumb|left|upright=.5]] | [[Image:FkAzFtQWIAExhDl.jpeg|link=https://globalbiodata.org/open-letter-campaign/|thumb|left|upright=.5]] | ||
| − | The [https://globalbiodata.org/ Global Biodata Coalition] has an open letter campaign to show support for sustainable funding for biodata resources. Please take a minute to read and sign- the form takes only seconds to fill out. Everyone- including students and postdocs, bioinformaticians and curators, PIs and directors - is invited to join those of us that have already added our signatures in support of the GBC. [https://globalbiodata.org/open-letter-campaign/ Find more information and sign at the GBC site]. | + | The [https://globalbiodata.org/ Global Biodata Coalition] has an open letter campaign to show support for sustainable funding for biodata resources. |
| − | + | ||
| + | Please take a minute to read and sign- the form takes only seconds to fill out. Everyone - including students and postdocs, bioinformaticians and curators, PIs and directors - is invited to join those of us that have already added our signatures in support of the GBC. [https://globalbiodata.org/open-letter-campaign/ Find more information and sign at the GBC site]. | ||
<br> | <br> | ||
<br> | <br> | ||
==Community wiki shutting down== | ==Community wiki shutting down== | ||
| − | As another casualty of decreased funding, SGD will no longer | + | As another casualty of decreased funding, SGD will no longer provide the Community Wiki. Some of the resources hosted on the wiki have been moved to our [https://sites.google.com/view/yeastgenome-help/sgd-general-help Help] pages, the [https://community.alliancegenome.org/c/model-organism-yeast/8 Alliance Community Forum], or remain available though the [http://sgd-archive.yeastgenome.org SGD downloads] site. We thank all our past and current Community Wiki contributors, and invite the yeast community to join SGD on the Alliance Community forum. |
==SGD’s new GENETICS publication== | ==SGD’s new GENETICS publication== | ||
| − | [[File:geneticslogo.png|link=https:// | + | [[File:geneticslogo.png|link=https://academic.oup.com/genetics/advance-article-abstract/doi/10.1093/genetics/iyae185/7895713|thumb|left|upright=.6]] |
[https://www.yeastgenome.org/reference/S000377741 ''Saccharomyces'' Genome Database: Advances in Genome Annotation, Expanded Biochemical Pathways, and Other Key Enhancements] has now been published in GENETICS and is available as an accepted manuscript. Check out the most recent updates at SGD, including the two most recent reference genome annotation updates, expanded biochemical pathways representation, changes to SGD search and data files, and other enhancements to the SGD website and user interface. | [https://www.yeastgenome.org/reference/S000377741 ''Saccharomyces'' Genome Database: Advances in Genome Annotation, Expanded Biochemical Pathways, and Other Key Enhancements] has now been published in GENETICS and is available as an accepted manuscript. Check out the most recent updates at SGD, including the two most recent reference genome annotation updates, expanded biochemical pathways representation, changes to SGD search and data files, and other enhancements to the SGD website and user interface. | ||
| − | ==microPublications - | + | ==microPublications - Latest yeast papers== |
[[Image:MicroPub.png|link=https://www.micropublication.org/|thumb|right|upright=.4]] | [[Image:MicroPub.png|link=https://www.micropublication.org/|thumb|right|upright=.4]] | ||
| Line 44: | Line 45: | ||
Latest [https://www.micropublication.org/journals/biology/species/s-cerevisiae yeast microPublications]: | Latest [https://www.micropublication.org/journals/biology/species/s-cerevisiae yeast microPublications]: | ||
| − | * [https://www.yeastgenome.org/reference/ | + | * [https://www.yeastgenome.org/reference/S000377792 Beard JS, Francis LK, Forrest RC, Kalinowski A, Parks JC, Griffin WH, Tackett CL, Duina AA (2024)] Trapping of yFACT at 3' ends of genes is not a universal characteristic of yeast versions of Bryant-Li-Bhoj syndrome histone H3 mutants. MicroPubl Biol 2024 |
| + | * [https://www.yeastgenome.org/reference/S000378016 Di Terlizzi M, Liberi G, Pellicioli A (2024)] Separation of function mutants underline multiple roles of the Srs2 helicase/translocase in break-induced replication in Saccharomyces cerevisiae. MicroPubl Biol 2024 | ||
| + | * [https://www.yeastgenome.org/reference/S000350461 Hill JM, Pedersen RT, Drubin DG (2024)] Myosin-I's motor and actin assembly activation activities are modular and separable in budding yeast clathrin-mediated endocytosis. MicroPubl Biol 2024 | ||
| + | * [https://www.yeastgenome.org/reference/S000375284 Miller JM, Tragesser-Tiña ME, Turk SM, Rubenstein EM (2024)] Loss of transcriptional regulator of phospholipid biosynthesis alters post-translational modification of Sec61 translocon beta subunit Sbh1 in ''Saccharomyces cerevisiae''. MicroPubl Biol 2024 | ||
* [https://www.yeastgenome.org/reference/S000376719 Mucelli X, Huang LS (2024)] Naming internal insertion alleles created using CRISPR in ''Saccharomyces cerevisiae''. MicroPubl Biol 2024 | * [https://www.yeastgenome.org/reference/S000376719 Mucelli X, Huang LS (2024)] Naming internal insertion alleles created using CRISPR in ''Saccharomyces cerevisiae''. MicroPubl Biol 2024 | ||
* [https://www.yeastgenome.org/reference/S000375481 Owutey SL, Procuniar KA, Akoto E, Davis JC, Vachon RM, O'Malley LF, Schneider HO, Smaldino PJ, True JD, Kalinski AL, Rubenstein EM (2024)] Endoplasmic reticulum and inner nuclear membrane ubiquitin-conjugating enzymes Ubc6 and Ubc7 confer resistance to hygromycin B in ''Saccharomyces cerevisiae''. MicroPubl Biol 2024 | * [https://www.yeastgenome.org/reference/S000375481 Owutey SL, Procuniar KA, Akoto E, Davis JC, Vachon RM, O'Malley LF, Schneider HO, Smaldino PJ, True JD, Kalinski AL, Rubenstein EM (2024)] Endoplasmic reticulum and inner nuclear membrane ubiquitin-conjugating enzymes Ubc6 and Ubc7 confer resistance to hygromycin B in ''Saccharomyces cerevisiae''. MicroPubl Biol 2024 | ||
| − | * [https://www.yeastgenome.org/reference/ | + | * [https://www.yeastgenome.org/reference/S000350355 Pinto J, Tavakolian N, Li CB, Stelkens R (2024)] The relationship between cell density and cell count differs among ''Saccharomyces'' yeast species. MicroPubl Biol 2024 |
* [https://www.yeastgenome.org/reference/S000377291 Ramakrishnan P, Keeney J (2024)] The yeast gene ECM9 regulates cell wall maintenance and cell division in stress conditions. MicroPubl Biol 2024 | * [https://www.yeastgenome.org/reference/S000377291 Ramakrishnan P, Keeney J (2024)] The yeast gene ECM9 regulates cell wall maintenance and cell division in stress conditions. MicroPubl Biol 2024 | ||
| − | |||
| − | |||
All yeast microPublications can be found in [https://www.yeastgenome.org/search?category=reference&journal=microPublication.%20Biology&page=0&q= SGD]. | All yeast microPublications can be found in [https://www.yeastgenome.org/search?category=reference&journal=microPublication.%20Biology&page=0&q= SGD]. | ||
| Line 59: | Line 61: | ||
The 7.4.0 release includes ~2.5 million variants of clinical significance from ClinVar. | The 7.4.0 release includes ~2.5 million variants of clinical significance from ClinVar. | ||
| − | The Alliance | + | The Alliance removed 400 million human variants from the search because their inclusion was hampering site performance. The removed variants are mostly of unknown or uncertain clinical significance. The Alliance has instead included over 2.5 million human variants of known clinical significance from ClinVar. This should speed up the performance of the search function and improve site stability. The Alliance will address inclusion of the other high throughput human variants in the future. More detailed information about the 7.4 Alliance release can be found in the [https://www.alliancegenome.org/release-notes release notes]. |
==Give a Gift / Support SGD== | ==Give a Gift / Support SGD== | ||
| Line 92: | Line 94: | ||
==Happy Holidays from SGD!== | ==Happy Holidays from SGD!== | ||
[[File:SnowShmoo.png|thumb|left|upright=.25|link=http://www.yeastgenome.org]] | [[File:SnowShmoo.png|thumb|left|upright=.25|link=http://www.yeastgenome.org]] | ||
| − | We want to take this opportunity to wish you and your family, friends, and lab mates the best during the upcoming holidays. Stanford University will be closed for two weeks starting December 23rd, reopening on January 6th, | + | We want to take this opportunity to wish you and your family, friends, and lab mates the best during the upcoming holidays. Stanford University will be closed for two weeks starting December 23rd, reopening on January 6th, 2025. Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will resume responding to user requests and questions in the new year. |
Latest revision as of 10:04, 13 December 2024
About this newsletter:
This is the December 2024 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features from SGD and to foster communication within the yeast community.
Contents
- 1 AlphaFold protein structures now on SGD protein pages
- 2 YeastMine data available in AllianceMine
- 3 Help maintain your genomic databases! Sign open letter supporting funding for biodata resources
- 4 Community wiki shutting down
- 5 SGD’s new GENETICS publication
- 6 microPublications - Latest yeast papers
- 7 Alliance of Genome Resources - Latest Release 7.4
- 8 Give a Gift / Support SGD
- 9 Upcoming conferences and courses
- 10 Happy Holidays from SGD!
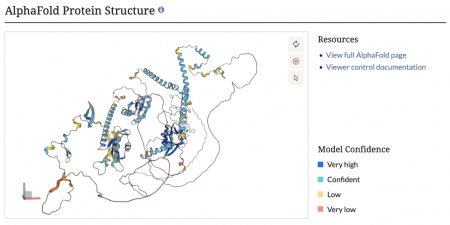
AlphaFold protein structures now on SGD protein pages
We are thrilled to announce that we have now integrated AlphaFold protein structures into our protein pages! This cutting-edge addition provides detailed, high-accuracy 3D models of protein structures, offering invaluable insights into protein function and interactions. Researchers can now explore these comprehensive structural predictions directly within SGD, facilitating advanced studies in molecular biology and bioinformatics. Dive into the new AlphaFold protein structures and elevate your research with this powerful tool!
AlphaFold, developed by DeepMind, is an AI program that accurately predicts protein structures from amino acid sequences, enabling visualization of protein conformations.
The predicted structures can be accessed through the Protein Data Bank (PDB) and AlphaFold Protein Structure Database. Thanks to Kim Rutherford and Val Wood of Pombase for tips about adding AlphaFold structures to SGD.
YeastMine data available in AllianceMine
Here at SGD we provide high-quality curated genomic, genetic, and molecular information on the genes and gene products of the budding yeast Saccharomyces cerevisiae. In 2011, SGD implemented InterMine, an open-source data warehouse system with a sophisticated querying interface, to better meet the complex and diverse needs of researchers searching and comparing data, resulting in the creation of YeastMine.
YeastMine is a multifaceted search and retrieval environment that provides access to diverse data types. Searches can be initiated with a list of genes, a list of Gene Ontology terms, or lists of many other data types. The results from queries can be combined for further analysis and saved or downloaded in customizable file formats. Queries themselves can be customized by modifying predefined templates or by creating a new template to access a combination of specific data types.
In July 2024, YeastMine was discontinued due to ongoing cuts in funding at SGD. However, we have moved the YeastMine data into AllianceMine, hosted by the Alliance of Genome Resources, of which SGD is a founding member. You can even access lists like “ALL_Verified_Uncharacterized_Dubious_ORFs” through the AllianceMine Lists just as you could in the previous versions of YeastMine. Ensure any bookmarks to YeastMine have been updated to match the new URL: https://www.alliancegenome.org/bluegenes/alliancemine. User documentation for the new YeastMine interface is available from InterMine.
Help maintain your genomic databases! Sign open letter supporting funding for biodata resources
The Global Biodata Coalition has an open letter campaign to show support for sustainable funding for biodata resources.
Please take a minute to read and sign- the form takes only seconds to fill out. Everyone - including students and postdocs, bioinformaticians and curators, PIs and directors - is invited to join those of us that have already added our signatures in support of the GBC. Find more information and sign at the GBC site.
Community wiki shutting down
As another casualty of decreased funding, SGD will no longer provide the Community Wiki. Some of the resources hosted on the wiki have been moved to our Help pages, the Alliance Community Forum, or remain available though the SGD downloads site. We thank all our past and current Community Wiki contributors, and invite the yeast community to join SGD on the Alliance Community forum.
SGD’s new GENETICS publication
Saccharomyces Genome Database: Advances in Genome Annotation, Expanded Biochemical Pathways, and Other Key Enhancements has now been published in GENETICS and is available as an accepted manuscript. Check out the most recent updates at SGD, including the two most recent reference genome annotation updates, expanded biochemical pathways representation, changes to SGD search and data files, and other enhancements to the SGD website and user interface.
microPublications - Latest yeast papers
microPublication Biology is part of the emerging genre of rapidly-published research communications. microPublications publishes brief, novel findings, negative and/or reproduced results, and results which may initially lack a broader scientific narrative. Each article is peer-reviewed, assigned a DOI, and indexed through PubMed and PubMedCentral.
Consider microPubublications when you have a result that doesn't necessarily fit into a larger story, but will be of value to others.
Latest yeast microPublications:
- Beard JS, Francis LK, Forrest RC, Kalinowski A, Parks JC, Griffin WH, Tackett CL, Duina AA (2024) Trapping of yFACT at 3' ends of genes is not a universal characteristic of yeast versions of Bryant-Li-Bhoj syndrome histone H3 mutants. MicroPubl Biol 2024
- Di Terlizzi M, Liberi G, Pellicioli A (2024) Separation of function mutants underline multiple roles of the Srs2 helicase/translocase in break-induced replication in Saccharomyces cerevisiae. MicroPubl Biol 2024
- Hill JM, Pedersen RT, Drubin DG (2024) Myosin-I's motor and actin assembly activation activities are modular and separable in budding yeast clathrin-mediated endocytosis. MicroPubl Biol 2024
- Miller JM, Tragesser-Tiña ME, Turk SM, Rubenstein EM (2024) Loss of transcriptional regulator of phospholipid biosynthesis alters post-translational modification of Sec61 translocon beta subunit Sbh1 in Saccharomyces cerevisiae. MicroPubl Biol 2024
- Mucelli X, Huang LS (2024) Naming internal insertion alleles created using CRISPR in Saccharomyces cerevisiae. MicroPubl Biol 2024
- Owutey SL, Procuniar KA, Akoto E, Davis JC, Vachon RM, O'Malley LF, Schneider HO, Smaldino PJ, True JD, Kalinski AL, Rubenstein EM (2024) Endoplasmic reticulum and inner nuclear membrane ubiquitin-conjugating enzymes Ubc6 and Ubc7 confer resistance to hygromycin B in Saccharomyces cerevisiae. MicroPubl Biol 2024
- Pinto J, Tavakolian N, Li CB, Stelkens R (2024) The relationship between cell density and cell count differs among Saccharomyces yeast species. MicroPubl Biol 2024
- Ramakrishnan P, Keeney J (2024) The yeast gene ECM9 regulates cell wall maintenance and cell division in stress conditions. MicroPubl Biol 2024
All yeast microPublications can be found in SGD.
Alliance of Genome Resources - Latest Release 7.4
The Alliance of Genome Resources, a collaborative effort between SGD and other model organism databases (MODs), released version 7.4 in October 2024.
The 7.4.0 release includes ~2.5 million variants of clinical significance from ClinVar. The Alliance removed 400 million human variants from the search because their inclusion was hampering site performance. The removed variants are mostly of unknown or uncertain clinical significance. The Alliance has instead included over 2.5 million human variants of known clinical significance from ClinVar. This should speed up the performance of the search function and improve site stability. The Alliance will address inclusion of the other high throughput human variants in the future. More detailed information about the 7.4 Alliance release can be found in the release notes.
Give a Gift / Support SGD
Budget cuts from NIH continue to strain SGD's finances. Despite our efforts at reducing costs, we still have significant ongoing budgetary challenges. Donations are now critical for our work to continue.
Your generous gift to SGD will help us to continue providing essential information for your research and teaching efforts.
To contribute, please make checks payable to Stanford University, noting that "the funds should be used to support the Saccharomyces Genome Database project, under the direction of Drs. Sherlock and Cherry in the Department of Genetics, Stanford University. Account : GHJKO, Genetics : WAZC."
Thank you for your support!
Kindly send by mail to:
Development Services
PO Box 20466
Stanford, CA 94309
CONTACT US: sgd-helpdesk@lists.stanford.edu
Upcoming conferences and courses
- Yeast2025: 32nd International Conference on Yeast Genetics and Molecular Biology ICYGMB32
- July 21 to July 24, 2025
- Sorbonne University, Paris, France
- 38th International Specialized Symposium on Yeasts (ISSY38)
- September 01 to September 05, 2025
- Warsaw University, Warsaw, Poland
Happy Holidays from SGD!
We want to take this opportunity to wish you and your family, friends, and lab mates the best during the upcoming holidays. Stanford University will be closed for two weeks starting December 23rd, reopening on January 6th, 2025. Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will resume responding to user requests and questions in the new year.