Difference between revisions of "SGD Newsletter, Summer 2023"
(Created page with "Category: Newsletter '''About this newsletter:''' <br> This is the Summer 2023 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new fe...") |
|||
| (74 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Category: Newsletter]] | [[Category: Newsletter]] | ||
| − | '''About this newsletter:''' <br> This is the Summer 2023 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can view this [https://wiki.yeastgenome.org/index.php/SGD_Newsletter,_Summer_2023 newsletter] as well as previous newsletters, on | + | '''About this newsletter:''' <br> This is the Summer 2023 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can view this [https://wiki.yeastgenome.org/index.php/SGD_Newsletter,_Summer_2023 newsletter] as well as previous newsletters, on the SGD [https://wiki.yeastgenome.org/index.php/SGD_Newsletter_Archives Community Wiki]. |
| − | == | + | ==Biochemical Pathways added to SGD Search== |
| − | [[Image: | + | [[Image:pathwaysSearch.png |thumb|left|upright=1.25]] |
| − | + | Biochemical pathways have been added to SGD search in order to facilitate easy access to metabolic yeast pathway pages at [http://pathway.yeastgenome.org YeastPathways]. | |
| − | + | YeastPathways is a database of metabolic pathways and enzymes in the budding yeast ''Saccharomyces cerevisiae''. YeastPathways content is manually curated and maintained by the curation team at SGD, your model organism database for budding yeast. Check it out! 👀 http://yeastgenome.org/search?q=&category=pathway | |
| − | |||
| − | + | Manual curation of pathways is an ongoing process at SGD. We welcome feedback from the research community. Please feel free to [mailto:sgd-helpdesk@lists.stanford.edu contact us] with any questions or comments. | |
| − | + | </p><p> | |
| + | <br /> | ||
| − | + | ==GENETICS Knowledgebase and Database Resources== | |
| + | [[Image:GENETICSmodUpdates.png|link=https://www.yeastgenome.org/blog/genetics-knowledgebase-and-database-resources |thumb|left|upright=.75]] | ||
| − | The | + | The [https://academic.oup.com/genetics/issue/224/1 May 2023 issue] of [https://academic.oup.com/genetics GENETICS] features the second annual collection of Model Organism Database articles. |
| − | + | Scientists from [https://www.alliancegenome.org Alliance of Genome Resources] member groups [https://www.yeastgenome.org SGD], [https://www.alliancegenome.org/members/rgd RGD], [https://www.alliancegenome.org/members/zfin ZFIN], [https://www.alliancegenome.org/members/goc Gene Ontology], and [https://www.xenbase.org/entry/ Xenbase] have provided updates on recent activities and innovations. | |
| + | |||
| + | Be sure to browse the issue and get acquainted with these excellent Knowledgebase and Database Resource papers at GENETICS. | ||
<br><br> | <br><br> | ||
| + | *SGD https://doi.org/10.1093/genetics/iyac191 | ||
| + | *RGD https://doi.org/10.1093/genetics/iyad042 | ||
| + | *ZFIN https://doi.org/10.1093/genetics/iyad032 | ||
| + | *Gene Ontology (GO) https://doi.org/10.1093/genetics/iyad031 | ||
| + | *Xenbase https://doi.org/10.1093/genetics/iyad018 | ||
| + | </p><p> | ||
| + | <br /> | ||
| − | == | + | ==SGD designated "Global Core Biodata Resource"== |
| − | [[ | + | [[Image:FkAzFtQWIAExhDl.jpeg|link=https://www.yeastgenome.org/blog/global-biodata-coalition-selects-sgd-in-first-set-of-global-core-biodata-resources |thumb|left|upright=.75]] |
| − | We | + | We are proud that SGD has been included in the first list of [https://globalbiodata.org/scientific-activities/global-core-biodata-resources/ Global Core Biodata Resources] (GCBRs) announced last December by the [https://globalbiodata.org/ Global Biodata Coalition] (GBC)! This collection of 37 resources comprises deposition databases which archive and preserve primary research data, and knowledgebases, such as SGD, that add value to research data through expert curation and annotation. The list is meant to highlight those data resources whose long term funding and sustainability is critical to life science and biomedical research worldwide. |
| − | + | GCBRs represent the most crucial resources within the global life science data community. SGD’s selection as a key global data resource recognizes that SGD is essential to the global research endeavor. | |
| + | For more information regarding the Global Biodata Coalition, including a link to the full list of selected core biodata resources, please see the full press release from the GBC. | ||
| + | </p><p> | ||
| + | <br /> | ||
| + | ==2D RNA structures from RNAcentral== | ||
| + | [[Image:RNAcentral.png|link=https://www.yeastgenome.org/blog/2d-rna-structures-from-rnacentral |thumb|right|upright=.4]] | ||
| + | SGD recently updated our RNA pages to add secondary structures provided by [https://rnacentral.org/ RNAcentral] and generated by [https://github.com/RNAcentral/r2dt R2DT]. | ||
| − | + | Thumbnails and linkouts to RNAcentral via RNAcentral IDs are shown on the [https://www.yeastgenome.org/locus/YNCQ0010W Summary] and Sequence pages. Interactive secondary structure viewers are available on the [https://www.yeastgenome.org/locus/YNCQ0010W/sequence#secondary_structure Sequence] pages. | |
| − | <br><br> | + | |
| + | Take the pages for a spin! For more information about the structures, please see the [https://rnacentral.org/help/secondary-structure Help page] at RNAcentral. | ||
| + | </p><p> | ||
| + | <br /> | ||
| + | </p><p> | ||
| + | <br /> | ||
| − | == | + | ==CoSMoS.c. - Conserved Sequence Motif in ''Saccharomyces cerevisiae''== |
| − | [[Image: | + | [[Image:cosmosc.png|thumb|left|upright=.5]] |
| − | + | The new website [https://shiny-server-dept-yeast-cosmos.apps.cloudapps.unc.edu CoSMoS.c.] - Conserved Sequence Motif in ''Saccharomyces cerevisiae'' - may be of interest to investigators who study protein modifications in budding yeast. The new web-based search algorithm scores conservation of amino acid sequences based on whole-genome sequencing of 1000+ wild and domesticated yeast isolates. | |
| − | + | In the [https://www.yeastgenome.org/reference/36933807 recent publication describing the method], Li and Dohlman examined each of the 550 pairs of duplicated genes in ''S. cerevisiae'', integrating 38,000+ documented post-translational modifications (PTMs), and 30,000+ reported interactions between protein kinases and substrates (all obtained from [https://www.yeastgenome.org SGD]!). More than 3,500 instances were identified where only one of two paralogous proteins undergoes a PTM despite having retained the same amino acid residue in both. Li and Dohlman found that the most common modifications – phosphorylation, ubiquitylation and acylation, but not N-glycosylation – occur in regions of high sequence conservation. The analysis indicates that differences in PTMs may be an important source of protein neo- or sub-functionalization, and that such differences likely account for the retention of closely related enzymes throughout evolution. | |
| − | + | You can find links to [https://shiny-server-dept-yeast-cosmos.apps.cloudapps.unc.edu CoSMoS.c.] at the bottom of your favorite protein page at SGD, in the Resources section under Post-translational Modifications. | |
| + | </p><p> | ||
| + | <br /> | ||
==microPublications - latest yeast papers== | ==microPublications - latest yeast papers== | ||
[[Image:MicroPub.png|link=https://www.micropublication.org/|thumb|right|upright=.4]] | [[Image:MicroPub.png|link=https://www.micropublication.org/|thumb|right|upright=.4]] | ||
| − | microPublication is part of the emerging genre of rapidly-published research communications. We are seeing a strong set of microPublications come through the database and are glad for this venue to publish brief, novel findings, negative and/or reproduced results, and results which may lack a broader scientific narrative. Each article is peer-reviewed, assigned a DOI, and indexed through PubMed and PubMedCentral. | + | [https://www.micropublication.org microPublication Biology] is part of the emerging genre of rapidly-published research communications. We are seeing a strong set of microPublications come through the database and are glad for this venue to publish brief, novel findings, negative and/or reproduced results, and results which may initially lack a broader scientific narrative. Each article is peer-reviewed, assigned a DOI, and indexed through PubMed and PubMedCentral. |
| + | Consider [https://www.micropublication.org/journals/biology/species/s-cerevisiae microPubublications] when you have a result that doesn't necessarily fit into a larger story, but will be of value to others. | ||
| − | + | Latest yeast microPublications: | |
| + | |||
| + | *[https://www.yeastgenome.org/reference/S000342338 Daraghmi MM, et al. (2023)] Macro-ER-phagy receptors Atg39p and Atg40p confer resistance to aminoglycoside hygromycin B in S. cerevisiae. MicroPubl Biol 2023 | ||
| + | *[https://www.yeastgenome.org/reference/S000342708 Domeni Zali G and Moriel-Carretero M (2023)] Auxin alone provokes retention of ASH1 mRNA in Saccharomyces cerevisiae mother cells. MicroPubl Biol 2023 | ||
| + | *[https://www.yeastgenome.org/reference/S000343723 Hiestand L, et al. (2023)] Chemical Genetics Screen of EVP4593 Sensitivity in Budding Yeast Identifies Effects on Mitochondrial Structure and Function. MicroPubl Biol 2023 | ||
| + | *[https://www.yeastgenome.org/reference/S000342340 Liu L, et al. (2023)] A role for ion homeostasis in yeast ionic liquid tolerance. MicroPubl Biol 2023 | ||
| + | *[https://www.yeastgenome.org/reference/S000342029 Longan ER, et al. (2023)] ADATscan - A flexible tool for scanning exomes for wobble inosine-dependent codons reveals a neurological bias for genes enriched in such codons in humans and mice. MicroPubl Biol 2023 | ||
| + | *[https://www.yeastgenome.org/reference/S000342948 Miles S, et al. (2023)] BY4741 cannot enter quiescence from rich medium. MicroPubl Biol 2023 | ||
| + | *[https://www.yeastgenome.org/reference/S000342432 Moresi NG, et al. (2023)] Caffeine-tolerant mutations selected through an at-home yeast experimental evolution teaching lab. MicroPubl Biol 2023 | ||
| + | *[https://www.yeastgenome.org/reference/S000342286 Shortt C, et al. (2023)] A simple and accessible CRISPR genome editing laboratory exercise using yeast. MicroPubl Biol 2023 | ||
| + | |||
| + | All yeast microPublications can be found in [https://www.yeastgenome.org/search?category=reference&journal=microPublication.%20Biology&page=0&q= SGD]. | ||
| + | </p><p> | ||
| + | <br /> | ||
| + | |||
| + | ==Alliance of Genome Resources - Releases 5.3 & 5.4== | ||
| + | [[Image:alliance_logo.png|link=https://www.alliancegenome.org/ |thumb|left|upright=.725]] | ||
| + | The [https://www.alliancegenome.org/ Alliance of Genome Resources], a collaborative effort from SGD and other model organism databases (MOD), released version 5.3 last October and [https://www.alliancegenome.org/release-notes version 5.4] this past April. | ||
| + | '''Version 5.3''' saw the addition of ''Xenopus'' data from Xenbase. These amphibians are an important tetrapod model, spanning the evolutionary gap between the mammalian (human, mouse, and rat) and zebrafish data already included in the Alliance. ''Xenopus'' have long been used in developmental and cell biology, and two species are widely studied. The African clawed frog, ''X. laevis'' is an allotetraploid (2n = 36) of hybrid origin. The resulting ''X. laevis'' genome has a set of ‘long’ and ‘short’ chromosomes and gene symbols are therefore appended with ‘.L’ or ‘.S’ denoting on which chromosome pair they reside. The second ''Xenopus'' species, the Western clawed frog ''X. tropicalis'', is a conventional diploid (2n=20), and is increasingly used in human disease modeling. | ||
| − | + | Data for both ''Xenopus'' species are organized on individual gene pages that feature data such as gene descriptions, orthology to human and other species, disease associations for orthologs, an expression ribbon view, and a Sequence Feature Viewer. | |
| − | + | '''Version 5.4''' provides new Alliance SimpleMine and Facebook URLs: | |
| − | |||
| − | |||
| − | * | + | *SimpleMine allows basic batch searches of Alliance data: https://www.alliancegenome.org/agr_simplemine.cgi |
| − | * | + | *The Alliance of Genome Resources has a new Facebook page: https://www.facebook.com/AllianceOfGenomeResources |
| + | </p><p> | ||
| + | <br /> | ||
==Upcoming Conferences and Courses== | ==Upcoming Conferences and Courses== | ||
| + | *[https://genomic.social/@DunhamLab/110042364409737514/ Pacific Northwest Yeast Club] | ||
| + | **Western Washington University, Bellingham, WA | ||
| + | **July 20, 2023 | ||
| + | |||
*[https://meetings.cshl.edu/courses.aspx?course=C-YEAS&year=22 Yeast Genetics & Genomics] - modern and intensive laboratory course that teaches students the full repertoire of genetic and genomic approaches needed to dissect complex problems using the yeast ''Saccharomyces cerevisiae'' | *[https://meetings.cshl.edu/courses.aspx?course=C-YEAS&year=22 Yeast Genetics & Genomics] - modern and intensive laboratory course that teaches students the full repertoire of genetic and genomic approaches needed to dissect complex problems using the yeast ''Saccharomyces cerevisiae'' | ||
**Cold Spring Harbor Laboratory, NY | **Cold Spring Harbor Laboratory, NY | ||
| − | **July 26 - August 15, | + | **July 26 - August 15, 2023 |
| − | *[https:// | + | |
| − | ** | + | *[https://blogs.cornell.edu/gibneylab/nery-2023/ 2023 Northeast Regional Yeast Meeting (NERY 2023)] |
| − | **August | + | **Cornell University, Ithaca, NY |
| − | * | + | **July 27 to July 28, 2023 |
| − | ** | + | |
| − | ** | + | *[https://www.yeastflorence2023.com ICYGMB31 - 31st International Conference on Yeast Genetics and Molecular Biology ] |
| − | **October | + | **Florence, Italy |
| − | *Fungal | + | **August 20 to August 25, 2023 |
| − | ** | + | |
| − | ** | + | *[https://smyte-2023.org 38th Small Meeting on Yeast Transport and Energetics (SMYTE)] |
| + | **Blankenberge, Belgium | ||
| + | **September 13 to September 17, 2023 | ||
| + | |||
| + | *[https://www.csh-asia.org/?content/1352 CSH Asia: Yeast and Life Sciences] | ||
| + | **Kunibiki Messe (Shimane Prefectural Convention Center), MATSUE, JAPAN | ||
| + | **October 09 to October 13, 2023 | ||
| + | |||
| + | *[https://issy2023.com.au ISSY2023: Yeast Biotech 2.0] | ||
| + | **National Wine Centre of Australia, Adelaide SA, Australia | ||
| + | **November 27 to December 01, 2023 | ||
| + | |||
| + | *[https://genetics-gsa.org/tagc/ TAGC2024 The Allied Genetics Conference] | ||
| + | **National Harbor | Washington DC Metro Area | ||
| + | **March 05 to March 10, 2024 | ||
| + | |||
| + | *[https://genetics-gsa.org/fungal/ 32nd Fungal Genetics Conference] | ||
| + | **Asilomar Conference Grounds, Pacific Grove, CA | ||
| + | **March 12 to March 17, 2024 | ||
| + | |||
| + | *[https://easternsun.eventsair.com/2024-16th-international-congress-on-yeasts ICY2024: 16th International Congress on Yeasts] | ||
| + | **Cape Town International Convention Centre, Cape Town, South Africa | ||
| + | **September 29 to October 03, 2024 | ||
Latest revision as of 14:18, 22 June 2023
About this newsletter:
This is the Summer 2023 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can view this newsletter as well as previous newsletters, on the SGD Community Wiki.
Contents
- 1 Biochemical Pathways added to SGD Search
- 2 GENETICS Knowledgebase and Database Resources
- 3 SGD designated "Global Core Biodata Resource"
- 4 2D RNA structures from RNAcentral
- 5 CoSMoS.c. - Conserved Sequence Motif in Saccharomyces cerevisiae
- 6 microPublications - latest yeast papers
- 7 Alliance of Genome Resources - Releases 5.3 & 5.4
- 8 Upcoming Conferences and Courses
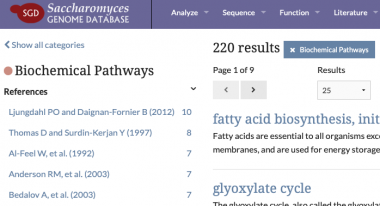
Biochemical Pathways added to SGD Search
Biochemical pathways have been added to SGD search in order to facilitate easy access to metabolic yeast pathway pages at YeastPathways.
YeastPathways is a database of metabolic pathways and enzymes in the budding yeast Saccharomyces cerevisiae. YeastPathways content is manually curated and maintained by the curation team at SGD, your model organism database for budding yeast. Check it out! 👀 http://yeastgenome.org/search?q=&category=pathway
Manual curation of pathways is an ongoing process at SGD. We welcome feedback from the research community. Please feel free to contact us with any questions or comments.
GENETICS Knowledgebase and Database Resources
The May 2023 issue of GENETICS features the second annual collection of Model Organism Database articles.
Scientists from Alliance of Genome Resources member groups SGD, RGD, ZFIN, Gene Ontology, and Xenbase have provided updates on recent activities and innovations.
Be sure to browse the issue and get acquainted with these excellent Knowledgebase and Database Resource papers at GENETICS.
- SGD https://doi.org/10.1093/genetics/iyac191
- RGD https://doi.org/10.1093/genetics/iyad042
- ZFIN https://doi.org/10.1093/genetics/iyad032
- Gene Ontology (GO) https://doi.org/10.1093/genetics/iyad031
- Xenbase https://doi.org/10.1093/genetics/iyad018
SGD designated "Global Core Biodata Resource"
We are proud that SGD has been included in the first list of Global Core Biodata Resources (GCBRs) announced last December by the Global Biodata Coalition (GBC)! This collection of 37 resources comprises deposition databases which archive and preserve primary research data, and knowledgebases, such as SGD, that add value to research data through expert curation and annotation. The list is meant to highlight those data resources whose long term funding and sustainability is critical to life science and biomedical research worldwide.
GCBRs represent the most crucial resources within the global life science data community. SGD’s selection as a key global data resource recognizes that SGD is essential to the global research endeavor.
For more information regarding the Global Biodata Coalition, including a link to the full list of selected core biodata resources, please see the full press release from the GBC.
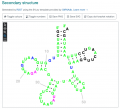
2D RNA structures from RNAcentral
SGD recently updated our RNA pages to add secondary structures provided by RNAcentral and generated by R2DT.
Thumbnails and linkouts to RNAcentral via RNAcentral IDs are shown on the Summary and Sequence pages. Interactive secondary structure viewers are available on the Sequence pages.
Take the pages for a spin! For more information about the structures, please see the Help page at RNAcentral.
CoSMoS.c. - Conserved Sequence Motif in Saccharomyces cerevisiae
The new website CoSMoS.c. - Conserved Sequence Motif in Saccharomyces cerevisiae - may be of interest to investigators who study protein modifications in budding yeast. The new web-based search algorithm scores conservation of amino acid sequences based on whole-genome sequencing of 1000+ wild and domesticated yeast isolates.
In the recent publication describing the method, Li and Dohlman examined each of the 550 pairs of duplicated genes in S. cerevisiae, integrating 38,000+ documented post-translational modifications (PTMs), and 30,000+ reported interactions between protein kinases and substrates (all obtained from SGD!). More than 3,500 instances were identified where only one of two paralogous proteins undergoes a PTM despite having retained the same amino acid residue in both. Li and Dohlman found that the most common modifications – phosphorylation, ubiquitylation and acylation, but not N-glycosylation – occur in regions of high sequence conservation. The analysis indicates that differences in PTMs may be an important source of protein neo- or sub-functionalization, and that such differences likely account for the retention of closely related enzymes throughout evolution.
You can find links to CoSMoS.c. at the bottom of your favorite protein page at SGD, in the Resources section under Post-translational Modifications.
microPublications - latest yeast papers
microPublication Biology is part of the emerging genre of rapidly-published research communications. We are seeing a strong set of microPublications come through the database and are glad for this venue to publish brief, novel findings, negative and/or reproduced results, and results which may initially lack a broader scientific narrative. Each article is peer-reviewed, assigned a DOI, and indexed through PubMed and PubMedCentral.
Consider microPubublications when you have a result that doesn't necessarily fit into a larger story, but will be of value to others.
Latest yeast microPublications:
- Daraghmi MM, et al. (2023) Macro-ER-phagy receptors Atg39p and Atg40p confer resistance to aminoglycoside hygromycin B in S. cerevisiae. MicroPubl Biol 2023
- Domeni Zali G and Moriel-Carretero M (2023) Auxin alone provokes retention of ASH1 mRNA in Saccharomyces cerevisiae mother cells. MicroPubl Biol 2023
- Hiestand L, et al. (2023) Chemical Genetics Screen of EVP4593 Sensitivity in Budding Yeast Identifies Effects on Mitochondrial Structure and Function. MicroPubl Biol 2023
- Liu L, et al. (2023) A role for ion homeostasis in yeast ionic liquid tolerance. MicroPubl Biol 2023
- Longan ER, et al. (2023) ADATscan - A flexible tool for scanning exomes for wobble inosine-dependent codons reveals a neurological bias for genes enriched in such codons in humans and mice. MicroPubl Biol 2023
- Miles S, et al. (2023) BY4741 cannot enter quiescence from rich medium. MicroPubl Biol 2023
- Moresi NG, et al. (2023) Caffeine-tolerant mutations selected through an at-home yeast experimental evolution teaching lab. MicroPubl Biol 2023
- Shortt C, et al. (2023) A simple and accessible CRISPR genome editing laboratory exercise using yeast. MicroPubl Biol 2023
All yeast microPublications can be found in SGD.
Alliance of Genome Resources - Releases 5.3 & 5.4
The Alliance of Genome Resources, a collaborative effort from SGD and other model organism databases (MOD), released version 5.3 last October and version 5.4 this past April.
Version 5.3 saw the addition of Xenopus data from Xenbase. These amphibians are an important tetrapod model, spanning the evolutionary gap between the mammalian (human, mouse, and rat) and zebrafish data already included in the Alliance. Xenopus have long been used in developmental and cell biology, and two species are widely studied. The African clawed frog, X. laevis is an allotetraploid (2n = 36) of hybrid origin. The resulting X. laevis genome has a set of ‘long’ and ‘short’ chromosomes and gene symbols are therefore appended with ‘.L’ or ‘.S’ denoting on which chromosome pair they reside. The second Xenopus species, the Western clawed frog X. tropicalis, is a conventional diploid (2n=20), and is increasingly used in human disease modeling.
Data for both Xenopus species are organized on individual gene pages that feature data such as gene descriptions, orthology to human and other species, disease associations for orthologs, an expression ribbon view, and a Sequence Feature Viewer.
Version 5.4 provides new Alliance SimpleMine and Facebook URLs:
- SimpleMine allows basic batch searches of Alliance data: https://www.alliancegenome.org/agr_simplemine.cgi
- The Alliance of Genome Resources has a new Facebook page: https://www.facebook.com/AllianceOfGenomeResources
Upcoming Conferences and Courses
- Pacific Northwest Yeast Club
- Western Washington University, Bellingham, WA
- July 20, 2023
- Yeast Genetics & Genomics - modern and intensive laboratory course that teaches students the full repertoire of genetic and genomic approaches needed to dissect complex problems using the yeast Saccharomyces cerevisiae
- Cold Spring Harbor Laboratory, NY
- July 26 - August 15, 2023
- 2023 Northeast Regional Yeast Meeting (NERY 2023)
- Cornell University, Ithaca, NY
- July 27 to July 28, 2023
- ICYGMB31 - 31st International Conference on Yeast Genetics and Molecular Biology
- Florence, Italy
- August 20 to August 25, 2023
- 38th Small Meeting on Yeast Transport and Energetics (SMYTE)
- Blankenberge, Belgium
- September 13 to September 17, 2023
- CSH Asia: Yeast and Life Sciences
- Kunibiki Messe (Shimane Prefectural Convention Center), MATSUE, JAPAN
- October 09 to October 13, 2023
- ISSY2023: Yeast Biotech 2.0
- National Wine Centre of Australia, Adelaide SA, Australia
- November 27 to December 01, 2023
- TAGC2024 The Allied Genetics Conference
- National Harbor | Washington DC Metro Area
- March 05 to March 10, 2024
- 32nd Fungal Genetics Conference
- Asilomar Conference Grounds, Pacific Grove, CA
- March 12 to March 17, 2024
- ICY2024: 16th International Congress on Yeasts
- Cape Town International Convention Centre, Cape Town, South Africa
- September 29 to October 03, 2024