Difference between revisions of "SGD Newsletter, Fall 2021"
(→Alliance of Genome Resources at https://www.alliancegenome.org/release-notes Version 4.1) |
(→Nomenclature Updates) |
||
| (80 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
[[Category: Newsletter]] | [[Category: Newsletter]] | ||
| − | '''About this newsletter:''' <br> This is the Fall 2021 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can view this [https://wiki.yeastgenome.org/index.php/SGD_Newsletter, | + | '''About this newsletter:''' <br> This is the Fall 2021 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can view this [https://wiki.yeastgenome.org/index.php/SGD_Newsletter,_Fall_2021 newsletter] as well as previous newsletters on our [https://wiki.yeastgenome.org/index.php/SGD_Newsletter_Archives Community Wiki]. |
| + | |||
| + | ==Protein Complex Page Updates== | ||
| + | [[File:Complex3.png|thumb|right|upright=1.3|link=https://www.yeastgenome.org/complex/CPX-1275]] | ||
| + | |||
| + | SGD has made recent updates to our protein complex pages to improve clarity and ease of use. The new pages for each complex will have the same format as gene pages, with tabs across the top for each category of information, including a [https://www.yeastgenome.org/complex/S000218162/ Summary] page, a [https://www.yeastgenome.org/complex/S000218162/go Gene Ontology] page, and a [https://www.yeastgenome.org/complex/S000218162/literature Literature] page. Just as we do for all of your favorite genes, Gene Ontology and Literature curation for complexes will be ongoing. | ||
| + | |||
| + | If you have any questions or feedback about the updates to our complex pages, please do not hesitate to [mailto:sgd-helpdesk@lists.stanford.edu contact us] at any time. | ||
| − | |||
==Nomenclature Updates== | ==Nomenclature Updates== | ||
| Line 24: | Line 30: | ||
|ARG56 | |ARG56 | ||
|- | |- | ||
| − | |[http://www.yeastgenome.org/locus/ | + | |[http://www.yeastgenome.org/locus/YBR208C YBR208C] |
|DUR1,2 | |DUR1,2 | ||
|DUR12 | |DUR12 | ||
| Line 30: | Line 36: | ||
|[http://www.yeastgenome.org/locus/YIL154C YIL154C] | |[http://www.yeastgenome.org/locus/YIL154C YIL154C] | ||
|IMP2' | |IMP2' | ||
| − | | | + | |IMP21 |
|- | |- | ||
|} | |} | ||
| − | ===New | + | ===New systematic nomenclature for yeast genes not in the reference genome=== |
For many years, a widely adopted systematic nomenclature has existed for yeast protein-coding genes, or ORFs, as many yeast researchers call them. Readers of the [https://wiki.yeastgenome.org/index.php/SGD_Newsletter,_Spring_2021 last SGD newsletter] will recall that, earlier this year, SGD adopted a new systematic nomenclature for the [https://www.yeastgenome.org/search?q=YNC&category=locus entire annotated complement of ncRNAs]. | For many years, a widely adopted systematic nomenclature has existed for yeast protein-coding genes, or ORFs, as many yeast researchers call them. Readers of the [https://wiki.yeastgenome.org/index.php/SGD_Newsletter,_Spring_2021 last SGD newsletter] will recall that, earlier this year, SGD adopted a new systematic nomenclature for the [https://www.yeastgenome.org/search?q=YNC&category=locus entire annotated complement of ncRNAs]. | ||
| Line 43: | Line 49: | ||
==New links to AlphaFold 3D Predicted Protein Structure Database== | ==New links to AlphaFold 3D Predicted Protein Structure Database== | ||
| − | https://alphafold.ebi.ac.uk/ | + | Would you like to see the shape of your protein? |
| + | [[File:Hog1_structure.png|thumb|left|upright=.5|link=https://alphafold.ebi.ac.uk/entry/P32485]] | ||
| − | [[ | + | SGD now contains links to [https://www.alphafold.ebi.ac.uk/ AlphaFold] in the Resources sections of the [https://www.yeastgenome.org/locus/S000004103#resources/ Summary], [https://www.yeastgenome.org/locus/hog1/protein#resources/ Protein], and [https://www.yeastgenome.org/locus/S000004103/homology#resources/ Homology] pages for every gene. |
| − | * | + | *The links through SGD give quick access to [https://www.embl.org EMBL]’s [https://www.ebi.ac.uk/ European Bioinformatics Institute] (EMBL-EBI), which offers a new, highly accurate tool for predicting protein structure with speed and clarity. |
*Given a peptide sequence for an uncharacterized protein, AlphaFold will model predicted domains and provide relative confidence levels for each portion of the prediction. | *Given a peptide sequence for an uncharacterized protein, AlphaFold will model predicted domains and provide relative confidence levels for each portion of the prediction. | ||
| − | *The predicted domains can then be compared to known protein structures (using a tool such as | + | *The predicted domains can then be compared to known protein structures (using a tool such as [https://www.ebi.ac.uk/msd-srv/ssm/ PDBeFold]) to seek matches to characterized protein families. |
*Whether or not a family is identified, the comparison will yield clues to protein function to help design the next experiments. | *Whether or not a family is identified, the comparison will yield clues to protein function to help design the next experiments. | ||
| − | ==YeastMine | + | ==DIOPT Orthologs and New Queries in YeastMine == |
| − | + | [[File:DIOPT-logo-integrative trans.png|thumb|left|upright=.5|link=https://www.flyrnai.org/diopt]] | |
| − | + | We recently replaced HomoloGene, Ensembl, TreeFam and PANTHER homology datasets in YeastMine with homology data from [https://www.flyrnai.org/diopt DIOPT (DRSC integrative ortholog prediction tool)]. DIOPT integrates orthology predictions from multiple sources, including HomoloGene, Ensembl, TreeFam, and PANTHER. Using the [https://yeastmine.yeastgenome.org/yeastmine/template.do?name=Gene_Homologs&scope=all Gene->Non-fungal and S. cerevisiae Homologs] pre-generated query, you can look for DIOPT homologs for a single or multiple yeast genes. The results table provides identifiers and standard names for the yeast and homologous genes, as well as organism and predictive score information. As with other YeastMine templates, results can be saved as lists and analyzed further.<br> | |
| − | |||
| + | Pre-generated [https://yeastmine.yeastgenome.org/yeastmine/template.do?name=Gene_OMIM_Disease&scope=all queries] for human homolog(s) of your favorite yeast gene and their corresponding disease associations remain largely unchanged. You can begin with your favorite human gene or disease keyword and retrieve the yeast counterparts of the relevant gene(s). As an example, you can search for the ''S. cerevisiae'' homologs of all human genes associated with disorders that contain the keyword “diabetes” ([https://yeastmine.yeastgenome.org/yeastmine/template.do?name=Disease_Human_Yeast&scope=all view search]). The results table provides identifiers and standard names for the yeast and human genes, OMIM gene and disease identifiers and name, as well as predictive algorithm sources and scores. | ||
| − | ==Alliance of Genome Resources - | + | ==Alliance of Genome Resources - Recent Release== |
| − | [[Image:alliance_logo.png|link=https://www.alliancegenome.org/ |thumb|left|upright=. | + | [[Image:alliance_logo.png|link=https://www.alliancegenome.org/ |thumb|left|upright=.5]] |
| − | The [https://www.alliancegenome.org/ Alliance of Genome Resources], a collaborative effort from SGD and other model organism databases (MOD), released version 4.1 | + | The [https://www.alliancegenome.org/ Alliance of Genome Resources], a collaborative effort from SGD and other model organism databases (MOD), released [https://www.alliancegenome.org/release-notes version 4.1] this past August. Notable improvements and new features included: |
| − | * | + | *Human and model organism high throughput (HTP) variant data |
| − | *Human variants are imported from Ensembl | + | **Human variants are imported from Ensembl |
| − | *Model organism HTP variants are submitted by Alliance members (FlyBase, RGD, SGD, Wormbase) or imported from EVA ( | + | **Model organism HTP variants are submitted by Alliance members (FlyBase, RGD, SGD, Wormbase) or imported from EVA (MGI and ZFIN). |
| − | *Added HTP variants to the Alleles and Variants table on gene pages (e.g. rat Lepr Gene page) and to the table on the Alleles and Variants Details page (e.g. rat Lepr Alleles and Variants Details. | + | **Added HTP variants to the Alleles and Variants table on gene pages (e.g. rat Lepr Gene page) and to the table on the Alleles and Variants Details page (e.g. rat Lepr Alleles and Variants Details. |
| − | *Created a report page for Human and model organism HTP variants (e.g. human variant rs1041354454). | + | **Created a report page for Human and model organism HTP variants (e.g. human variant rs1041354454). |
| − | *Expanded Allele Category in search to “Allele/Variant” and added a search for HTP variants | + | **Expanded Allele Category in search to “Allele/Variant” and added a search for HTP variants. |
| − | |||
*On Gene Pages, a new Pathways widget displays via tabs: | *On Gene Pages, a new Pathways widget displays via tabs: | ||
**Reactome models of pathways for human gene products as well as inferred pathways for model organism genes based on orthology to human genes. | **Reactome models of pathways for human gene products as well as inferred pathways for model organism genes based on orthology to human genes. | ||
**Reactome reactions for gene products (e.g. human TP53 Gene page) | **Reactome reactions for gene products (e.g. human TP53 Gene page) | ||
| − | *Gene Ontology Causal Activity Models (GO-CAMs). These provide a framework to represent a biological system by linking together multiple GO annotations. PMID:31548717 (e.g. worm nsy-1 Gene page). | + | **Gene Ontology Causal Activity Models (GO-CAMs). These provide a framework to represent a biological system by linking together multiple GO annotations. PMID:31548717 (e.g. worm nsy-1 Gene page). |
*Experimental conditions are include for Disease and Phenotype data in tables on Gene, Allele, and Disease pages (e.g. zebrafish scn1lab Gene page). | *Experimental conditions are include for Disease and Phenotype data in tables on Gene, Allele, and Disease pages (e.g. zebrafish scn1lab Gene page). | ||
*AllianceMine added Orthologs, and Allele and Variants (low throughput) data types to this release. You can now query for these data types via pre-made template queries. | *AllianceMine added Orthologs, and Allele and Variants (low throughput) data types to this release. You can now query for these data types via pre-made template queries. | ||
*The Alliance Community Forum is released. The Forum permits discussions across six model organism communities—flies, mice, yeast, rats, worms, and zebrafish. More details will follow. | *The Alliance Community Forum is released. The Forum permits discussions across six model organism communities—flies, mice, yeast, rats, worms, and zebrafish. More details will follow. | ||
| − | ==Upcoming Conferences== | + | ==Upcoming Conferences and Courses== |
*[https://genetics-gsa.org/fungal/ Fungal Genetics] - the premier meeting for the international community of fungal geneticists | *[https://genetics-gsa.org/fungal/ Fungal Genetics] - the premier meeting for the international community of fungal geneticists | ||
**Asilomar Conference Grounds, Pacific Grove, California (and Online) | **Asilomar Conference Grounds, Pacific Grove, California (and Online) | ||
| Line 86: | Line 92: | ||
**University of British Columbia, Vancouver | **University of British Columbia, Vancouver | ||
**July 12 - 16, 2022 | **July 12 - 16, 2022 | ||
| + | *[https://meetings.cshl.edu/courses.aspx?course=C-YEAS&year=22 CSHL Yeast Genetics & Genomics] - modern, intensive laboratory course that teaches students full repertoire of genetic and genomic approaches | ||
| + | **Cold Spring Harbor Laboratory, NY | ||
| + | **July 26 - August 15, 2022 | ||
*[https://genetics-gsa.org/yeast/ Yeast Genetics Meeting] - the premier meeting for students, postdoctoral scholars, research staff, and principal investigators studying various aspects of eukaryotic biology in yeast | *[https://genetics-gsa.org/yeast/ Yeast Genetics Meeting] - the premier meeting for students, postdoctoral scholars, research staff, and principal investigators studying various aspects of eukaryotic biology in yeast | ||
**University of California, Los Angeles | **University of California, Los Angeles | ||
| Line 101: | Line 110: | ||
==Happy Holidays from SGD!== | ==Happy Holidays from SGD!== | ||
| − | [[File:SnowShmoo.png|thumb|left|upright=.25]] | + | [[File:SnowShmoo.png|thumb|left|upright=.25|link=http://www.yeastgenome.org]] |
We know that 2021 has been another challenging year for everyone. Our thoughts go out to all those who have been impacted by recent events. We wish you and your family, friends, and lab mates the best during the upcoming holidays. | We know that 2021 has been another challenging year for everyone. Our thoughts go out to all those who have been impacted by recent events. We wish you and your family, friends, and lab mates the best during the upcoming holidays. | ||
| − | '''Stanford University will be closed for two weeks starting | + | '''Stanford University will be closed for two weeks starting December 20, and will reopen on January 3rd, 2022'''. Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will resume responding to user requests and questions in the new year. |
<br><br> | <br><br> | ||
Latest revision as of 08:42, 16 December 2021
About this newsletter:
This is the Fall 2021 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can view this newsletter as well as previous newsletters on our Community Wiki.
Contents
[hide]- 1 Protein Complex Page Updates
- 2 Nomenclature Updates
- 3 New links to AlphaFold 3D Predicted Protein Structure Database
- 4 DIOPT Orthologs and New Queries in YeastMine
- 5 Alliance of Genome Resources - Recent Release
- 6 Upcoming Conferences and Courses
- 7 Gene Ontology Consortium Fall 2021 Meeting
- 8 Happy Holidays from SGD!
Protein Complex Page Updates
SGD has made recent updates to our protein complex pages to improve clarity and ease of use. The new pages for each complex will have the same format as gene pages, with tabs across the top for each category of information, including a Summary page, a Gene Ontology page, and a Literature page. Just as we do for all of your favorite genes, Gene Ontology and Literature curation for complexes will be ongoing.
If you have any questions or feedback about the updates to our complex pages, please do not hesitate to contact us at any time.
Nomenclature Updates
Legacy gene names
SGD has long been the keeper of the official Saccharomyces cerevisiae gene nomenclature. Robert Mortimer handed over this responsibility to SGD in 1993 after maintaining the yeast genetic map and gene nomenclature for 30 years.
The accepted format for gene names in S. cerevisiae comprises three uppercase letters followed by a number. The letters typically signify a phrase (referred to as the "Name Description" in SGD) that provides information about a function, mutant phenotype, or process related to that gene, for example "ADE" for "ADEnine biosynthesis" or "CDC" for "Cell Division Cycle". Gene names for many types of chromosomal features follow this basic format regardless of the type of feature named, whether an ORF, a tRNA, another type of non-coding RNA, an ARS, or a genetic locus. Some S. cerevisiae gene names that pre-date the current nomenclature standards do not conform to this format, such as MRLP38, RPL1A, and OM45.
A few historical gene names predate both the nomenclature standards and the database, and were less computer-friendly than more recent gene names, due to the presence of punctuation. SGD recently updated these gene names to be consistent with current standards and to be more software-friendly by removing punctuation. The old names for these four genes have been retained as aliases.
| ORF | Old gene name | New gene name |
|---|---|---|
| YGL234W | ADE5,7 | ADE57 |
| YER069W | ARG5,6 | ARG56 |
| YBR208C | DUR1,2 | DUR12 |
| YIL154C | IMP2' | IMP21 |
New systematic nomenclature for yeast genes not in the reference genome
For many years, a widely adopted systematic nomenclature has existed for yeast protein-coding genes, or ORFs, as many yeast researchers call them. Readers of the last SGD newsletter will recall that, earlier this year, SGD adopted a new systematic nomenclature for the entire annotated complement of ncRNAs.
We have just put into place a new systematic nomenclature for S. cerevisiae genes that are not found in the reference genome of strain S288C ("non-reference" genes). This new systematic nomenclature is similar to, but distinct from, that used for ORFs and that used for ncRNAs. Non-reference genes are designated by a symbol consisting of three uppercase letters and a four-digit number, as follows: Y for "Yeast", SC for "Saccharomyces cerevisiae", and a four-digit number corresponding to the sequential order in which the gene was added to SGD. We currently have 55 of these genes in SGD, some of which are old favorites like MAL21/YSC0004 and MATA/YSC0046, while others are more recent additions like XDH1/YSC0051. Going forward, as evidence is published pointing to other S. cerevisiae genes not present in the S288C reference genome, they will be added to the annotation using the next sequential number available. We already have 15 more of these YSC0000 names reserved by researchers and awaiting publication.
If you have some non-reference genes for which these names would be appropriate, please let us know!
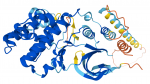
New links to AlphaFold 3D Predicted Protein Structure Database
Would you like to see the shape of your protein?
SGD now contains links to AlphaFold in the Resources sections of the Summary, Protein, and Homology pages for every gene.
- The links through SGD give quick access to EMBL’s European Bioinformatics Institute (EMBL-EBI), which offers a new, highly accurate tool for predicting protein structure with speed and clarity.
- Given a peptide sequence for an uncharacterized protein, AlphaFold will model predicted domains and provide relative confidence levels for each portion of the prediction.
- The predicted domains can then be compared to known protein structures (using a tool such as PDBeFold) to seek matches to characterized protein families.
- Whether or not a family is identified, the comparison will yield clues to protein function to help design the next experiments.
DIOPT Orthologs and New Queries in YeastMine
We recently replaced HomoloGene, Ensembl, TreeFam and PANTHER homology datasets in YeastMine with homology data from DIOPT (DRSC integrative ortholog prediction tool). DIOPT integrates orthology predictions from multiple sources, including HomoloGene, Ensembl, TreeFam, and PANTHER. Using the Gene->Non-fungal and S. cerevisiae Homologs pre-generated query, you can look for DIOPT homologs for a single or multiple yeast genes. The results table provides identifiers and standard names for the yeast and homologous genes, as well as organism and predictive score information. As with other YeastMine templates, results can be saved as lists and analyzed further.
Pre-generated queries for human homolog(s) of your favorite yeast gene and their corresponding disease associations remain largely unchanged. You can begin with your favorite human gene or disease keyword and retrieve the yeast counterparts of the relevant gene(s). As an example, you can search for the S. cerevisiae homologs of all human genes associated with disorders that contain the keyword “diabetes” (view search). The results table provides identifiers and standard names for the yeast and human genes, OMIM gene and disease identifiers and name, as well as predictive algorithm sources and scores.
Alliance of Genome Resources - Recent Release
The Alliance of Genome Resources, a collaborative effort from SGD and other model organism databases (MOD), released version 4.1 this past August. Notable improvements and new features included:
- Human and model organism high throughput (HTP) variant data
- Human variants are imported from Ensembl
- Model organism HTP variants are submitted by Alliance members (FlyBase, RGD, SGD, Wormbase) or imported from EVA (MGI and ZFIN).
- Added HTP variants to the Alleles and Variants table on gene pages (e.g. rat Lepr Gene page) and to the table on the Alleles and Variants Details page (e.g. rat Lepr Alleles and Variants Details.
- Created a report page for Human and model organism HTP variants (e.g. human variant rs1041354454).
- Expanded Allele Category in search to “Allele/Variant” and added a search for HTP variants.
- On Gene Pages, a new Pathways widget displays via tabs:
- Reactome models of pathways for human gene products as well as inferred pathways for model organism genes based on orthology to human genes.
- Reactome reactions for gene products (e.g. human TP53 Gene page)
- Gene Ontology Causal Activity Models (GO-CAMs). These provide a framework to represent a biological system by linking together multiple GO annotations. PMID:31548717 (e.g. worm nsy-1 Gene page).
- Experimental conditions are include for Disease and Phenotype data in tables on Gene, Allele, and Disease pages (e.g. zebrafish scn1lab Gene page).
- AllianceMine added Orthologs, and Allele and Variants (low throughput) data types to this release. You can now query for these data types via pre-made template queries.
- The Alliance Community Forum is released. The Forum permits discussions across six model organism communities—flies, mice, yeast, rats, worms, and zebrafish. More details will follow.
Upcoming Conferences and Courses
- Fungal Genetics - the premier meeting for the international community of fungal geneticists
- Asilomar Conference Grounds, Pacific Grove, California (and Online)
- March 15 - 20, 2022
- 36th International Specialised Symposium on Yeasts (ISSY36) - Yeast Sea to Sky - Yeast in the Genomics Era
- University of British Columbia, Vancouver
- July 12 - 16, 2022
- CSHL Yeast Genetics & Genomics - modern, intensive laboratory course that teaches students full repertoire of genetic and genomic approaches
- Cold Spring Harbor Laboratory, NY
- July 26 - August 15, 2022
- Yeast Genetics Meeting - the premier meeting for students, postdoctoral scholars, research staff, and principal investigators studying various aspects of eukaryotic biology in yeast
- University of California, Los Angeles
- August 17 – 21, 2022
Gene Ontology Consortium Fall 2021 Meeting
From October 12-14, SGD biocurators attended the Gene Ontology Consortium's Fall Meeting with participants from around the world. The goal of these meetings is to bring together data scientists with diverse backgrounds (curators, programmers, etc.) for lively discussions regarding how to better capture, curate, analyze, and serve data to researchers, educators, students, and other life science professionals. Our goal in participating in these meetings each year is to find ways to make SGD even better for you!
Discussion topics included, but were not limited to:
- LitSuggest - web-based system for biomedical literature recommendation and curation
- ECO, Evidence and Conclusions Ontology - terms used to describe types of evidence and assertion methods
- PAINT, Phylogenetic Annotation and INference Tool from PANTHER - orthology between reference genome genes and human disease genes
Happy Holidays from SGD!
We know that 2021 has been another challenging year for everyone. Our thoughts go out to all those who have been impacted by recent events. We wish you and your family, friends, and lab mates the best during the upcoming holidays.
Stanford University will be closed for two weeks starting December 20, and will reopen on January 3rd, 2022. Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will resume responding to user requests and questions in the new year.