Difference between revisions of "SGD Newsletter, Fall 2020"
Mjalexander (talk | contribs) |
Mjalexander (talk | contribs) (→Interaction Page Updates) |
||
| (173 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
'''About this newsletter:''' <br> | '''About this newsletter:''' <br> | ||
| − | This is the Fall 2020 issue of the SGD | + | This is the Fall 2020 issue of the SGD Newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. |
| − | You can | + | You can view this [https://wiki.yeastgenome.org/index.php/SGD_Newsletter,_Fall_2020 newsletter] as well as previous newsletters on our [https://wiki.yeastgenome.org/index.php/SGD_Newsletter_Archives Community Wiki]. |
| + | |||
==New SGD Allele Pages== | ==New SGD Allele Pages== | ||
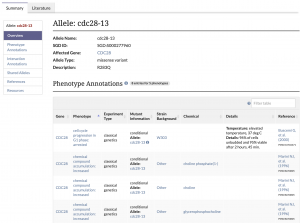
| − | [[File:allele_page.png|link=https://yeastgenome.org/allele/S000277751|thumb|left|upright=.8|The cdc28-13 allele page.]] | + | [[File:allele_page.png|link=https://yeastgenome.org/allele/S000277751|300px|thumb|left|upright=.8|The cdc28-13 allele page.]] |
| − | + | We are pleased to announce that SGD’s brand new Allele Pages are now available on our website. To navigate to an allele page, use the search bar to find a specific allele or enter a gene name and select an allele from the autocomplete list. Additionally, these pages can be accessed by clicking on the allele name in a gene’s Phenotype Annotation table. These pages are still being updated with new information as it becomes available. | |
The type of information that you can find on each allele page includes: | The type of information that you can find on each allele page includes: | ||
| − | + | *'''Allele Overview:''' General information about the allele, such as its name, the affected gene, the type of allele (e.g. missense), and a description of sequence change and/or domain mutated. | |
| − | + | *'''Phenotype and Interaction Annotations:''' Phenotype and Genetic Interaction Annotation tables for the allele. | |
| − | + | *'''Shared Alleles:''' A network diagram depicting shared phenotypes and interactions with other alleles. | |
If you are interested in viewing all alleles for a specific gene or would like to view a comprehensive list of the alleles that SGD currently has curated, you can use [https://yeastmine.yeastgenome.org/yeastmine/template.do?name=Gene_Alleles&scope=all this YeastMine template] with your customized parameters. | If you are interested in viewing all alleles for a specific gene or would like to view a comprehensive list of the alleles that SGD currently has curated, you can use [https://yeastmine.yeastgenome.org/yeastmine/template.do?name=Gene_Alleles&scope=all this YeastMine template] with your customized parameters. | ||
<br> | <br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
==Interaction Page Updates== | ==Interaction Page Updates== | ||
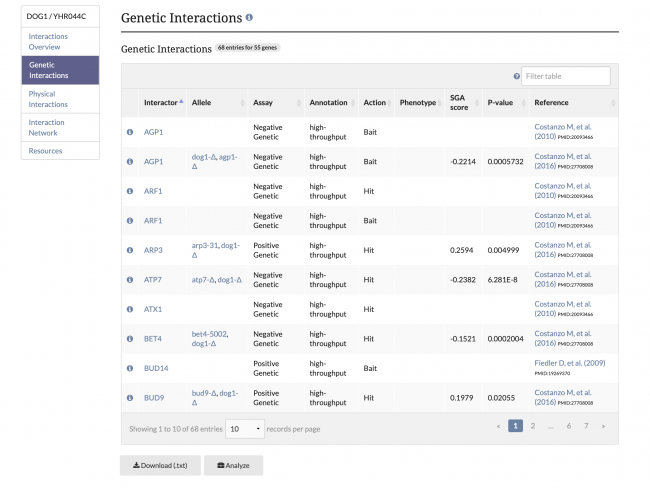
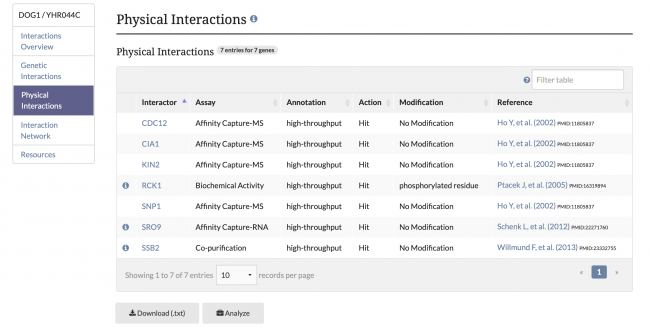
| − | [[File: | + | SGD has made recent updates to our Gene Interactions Page for improved clarity. Previously, genetic and physical interaction annotations were combined in one table, but now these annotations are recorded in separate annotation tables. The menu in the top left corner can be used to view and navigate to each section of the Interactions page. Additionally, alleles, SGA Scores, and P-values are now included for annotations from the global interactions paper by [https://www.yeastgenome.org/reference/S000185095 Costanzo M, et al. (2016)]. |
| + | <br> | ||
| + | |||
| + | |||
| + | [[File:genetic_interactions.png|650px|left|Image: 650 pixels]] [[File:physical_interactions.png|650px|Image: 650 pixels]] | ||
| + | |||
| + | ==Links on SGD gene pages to homolog gene pages at the Alliance of Genome Resources== | ||
| + | [[File:alliance_links.png|thumb|left|upright=.8|]] | ||
| + | [[File:alliance_links_2.png|thumb|right|upright=.8|]] | ||
| + | Previously, the 'Comparative Info' section on the Gene Summary page contained a link to the [https://www.alliancegenome.org Alliance of Genome Resources] if integrated model organism details were available. Now, users will instead find hexagonal buttons representing each model organism (human, mouse, rat, zebrafish, fly, and worm) for which there is homologous gene information at the Alliance of Genome Resources. Clicking on the link will immediately direct the user to the gene page for the selected model organism on the Alliance website. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | ==Alliance of Genome Resources at Version 3.2== | + | ==Alliance of Genome Resources at [https://www.alliancegenome.org/release-notes Version 3.2]== |
[[Image:alliance_logo.png|link=https://www.alliancegenome.org/ |thumb|left|upright=.7]] | [[Image:alliance_logo.png|link=https://www.alliancegenome.org/ |thumb|left|upright=.7]] | ||
The [https://www.alliancegenome.org/ Alliance of Genome Resources], a collaborative effort from SGD and other model organism databases (MOD), released version 3.2 in October. Notable improvements and new features include: | The [https://www.alliancegenome.org/ Alliance of Genome Resources], a collaborative effort from SGD and other model organism databases (MOD), released version 3.2 in October. Notable improvements and new features include: | ||
| Line 35: | Line 46: | ||
*"NOT" disease annotations are shown in Disease, Alleles and Variants, and Models sections when an anticipated disease annotation was not found. | *"NOT" disease annotations are shown in Disease, Alleles and Variants, and Models sections when an anticipated disease annotation was not found. | ||
*Enhanced variant data representation on Allele pages. | *Enhanced variant data representation on Allele pages. | ||
| − | *High-throughput profiling experiment metadata can be searched and will display in the search results as HTP Dataset Index | + | *High-throughput profiling experiment metadata can be searched and will display in the search results as HTP Dataset Index. |
| + | *The [https://www.alliancegenome.org/news/the-alliance-user-community-discussion-forum-was-released-on-october-14-2020 Alliance User Community discussion forum] has been released. | ||
| + | |||
| + | ==Jeremy Thorner Retiring== | ||
| + | [[File:jeremy_thorner.jpeg|150px|thumb|left|upright=1]] | ||
| + | Jeremy Thorner, a prolific yeast researcher and outstanding mentor, has announced that his lab is closing. His lab provided the following notice: | ||
| + | |||
| + | "After 47 years at the University of California, Berkeley, the laboratory of Professor Jeremy Thorner will be closing permanently, as of 30 June 2021. After that date, there will be no way to distribute any strains, plasmids, enzymes, or antibodies generated during the course of the studies on ''Saccharomyces cerevisiae'' conducted by the Thorner laboratory over those many years." | ||
| − | + | <br> | |
| − | |||
| − | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | |
| − | + | ||
| − | [[File: | + | |
| − | + | ==YeaZ system for microscopy images== | |
| − | + | [[File:sgdwiki-yeaz-screenshot.png|300px|thumb|left|upright=1]] | |
| − | + | [https://www.epfl.ch/labs/lpbs/data-and-software/ YeaZ] is a system for efficiently and accurately segmenting microscopy images of yeast cells. It contains a convolutional neural network, with an underlying training set of high-qual|ty segmented yeast images, as well as a graphical user interface and a web application to employ, test, and expand the system. The system contains a Python based application with graphical user interface available on [https://github.com/lpbsscientist/YeaZ-GUI GitHub], as well as standalone apps for both Windows and Mac based computers, and training sets. Additional information is available in the accompanying Nature Communications paper by [https://www.nature.com/articles/s41467-020-19557-4 Dietler et al., 2020]. | |
| − | + | ||
| − | + | YeaZ was created at École polytechnique fédérale de Lausanne (EPFL), Lausanne, Switzerland. Please contact [https://www.epfl.ch/labs/lpbs/professor-rahi/ Sahand Jamal Rahi] with questions, or ideas for improvements. | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==In Memoriam: Angelika Amon== | ||
| + | [[File:angelika_amon.jpeg|150px|thumb|left|upright=1]] | ||
| + | It was with great sadness that we learned that Angelika Amon, cell biologist and Professor of Biology at MIT and a member of the Koch Institute for Integrative Cancer Research, passed away on October 29, 2020. Angelika was widely known for her novel contributions to the field of cell biology and proliferation, with her research focusing on aneuploidy and the consequences of chromosome mis-segregation. At the beginning of Angelika's career, her work on yeast genetics led to the discovery that cyclins must break down completely before cells progress from mitosis to G1. | ||
| + | |||
| + | Her research has helped shape the current understanding of cell division, and her passion for genetics will live on through her students and colleagues who continue her work. Angelika was a treasured part of the genetics community and will be missed dearly. | ||
| + | |||
| + | The Genetics Society of America has featured [http://genestogenomes.org/in-memoriam-angelika-amon-a-brilliant-scientist-and-a-dear-friend/ a tribute piece in remembrance of Angelika Amon], written by her friend and colleague, Orna Cohen-Fix. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==Gene Ontology Consortium Meeting== | ||
| + | [[File:goc_group_photo.png|thumb|left|upright=1]] | ||
| + | From October 6th-8th, PI Mike Cherry, Senior Biocuration Scientist Edith Wong, Senior Biocuration Scientist Rob Nash, Senior Biocuration Scientist Marek Skrzypek, Biocuration Scientist Suzi Aleksander, and Associate Biocuration Scientist Micheal Alexander attended the Gene Ontology Consortium meeting. Suzi presented on SGD’s GO display and GO tools, and curators learned more about how other databases utilize GO. SGD Curators also participated in meaningful discussions about improving existing GO resources while also helping with the planning of new GO projects. | ||
| + | |||
| + | We would like to thank The Gene Ontology for facilitating this successful change to a virtual conference and holding an accessible, well received event for the entire model organism community! | ||
| + | |||
| + | |||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==Happy Holidays from SGD!== | |
| − | + | [[File:SnowShmoo.png|thumb|left|upright=.25]] | |
| − | + | We would like to take this opportunity to recognize that 2020 has brought many changes and challenges for everyone. Our thoughts go out to all those who have been impacted by the unprecedented events of this year. | |
| − | |||
| − | |||
| − | + | We wish you and your family, friends, and lab mates the best during the upcoming holidays. '''Stanford University will be closed for three weeks starting on December 14 and will reopen on January 4th, 2021'''. Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will resume responding to user requests and questions in the new year. | |
| − | + | <br><br> | |
| − | |||
Latest revision as of 14:06, 24 May 2021
About this newsletter:
This is the Fall 2020 issue of the SGD Newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community. You can view this newsletter as well as previous newsletters on our Community Wiki.
Contents
- 1 New SGD Allele Pages
- 2 Interaction Page Updates
- 3 Links on SGD gene pages to homolog gene pages at the Alliance of Genome Resources
- 4 Alliance of Genome Resources at Version 3.2
- 5 Jeremy Thorner Retiring
- 6 YeaZ system for microscopy images
- 7 In Memoriam: Angelika Amon
- 8 Gene Ontology Consortium Meeting
- 9 Happy Holidays from SGD!
New SGD Allele Pages
We are pleased to announce that SGD’s brand new Allele Pages are now available on our website. To navigate to an allele page, use the search bar to find a specific allele or enter a gene name and select an allele from the autocomplete list. Additionally, these pages can be accessed by clicking on the allele name in a gene’s Phenotype Annotation table. These pages are still being updated with new information as it becomes available. The type of information that you can find on each allele page includes:
- Allele Overview: General information about the allele, such as its name, the affected gene, the type of allele (e.g. missense), and a description of sequence change and/or domain mutated.
- Phenotype and Interaction Annotations: Phenotype and Genetic Interaction Annotation tables for the allele.
- Shared Alleles: A network diagram depicting shared phenotypes and interactions with other alleles.
If you are interested in viewing all alleles for a specific gene or would like to view a comprehensive list of the alleles that SGD currently has curated, you can use this YeastMine template with your customized parameters.
Interaction Page Updates
SGD has made recent updates to our Gene Interactions Page for improved clarity. Previously, genetic and physical interaction annotations were combined in one table, but now these annotations are recorded in separate annotation tables. The menu in the top left corner can be used to view and navigate to each section of the Interactions page. Additionally, alleles, SGA Scores, and P-values are now included for annotations from the global interactions paper by Costanzo M, et al. (2016).
Links on SGD gene pages to homolog gene pages at the Alliance of Genome Resources
Previously, the 'Comparative Info' section on the Gene Summary page contained a link to the Alliance of Genome Resources if integrated model organism details were available. Now, users will instead find hexagonal buttons representing each model organism (human, mouse, rat, zebrafish, fly, and worm) for which there is homologous gene information at the Alliance of Genome Resources. Clicking on the link will immediately direct the user to the gene page for the selected model organism on the Alliance website.
Alliance of Genome Resources at Version 3.2
The Alliance of Genome Resources, a collaborative effort from SGD and other model organism databases (MOD), released version 3.2 in October. Notable improvements and new features include:
- Gene pages now display transgenic allele data under the 'Transgenic Allele' section.
- Allele and Variant sections on Gene pages have improved formatting and are now downloadable.
- "NOT" disease annotations are shown in Disease, Alleles and Variants, and Models sections when an anticipated disease annotation was not found.
- Enhanced variant data representation on Allele pages.
- High-throughput profiling experiment metadata can be searched and will display in the search results as HTP Dataset Index.
- The Alliance User Community discussion forum has been released.
Jeremy Thorner Retiring
Jeremy Thorner, a prolific yeast researcher and outstanding mentor, has announced that his lab is closing. His lab provided the following notice:
"After 47 years at the University of California, Berkeley, the laboratory of Professor Jeremy Thorner will be closing permanently, as of 30 June 2021. After that date, there will be no way to distribute any strains, plasmids, enzymes, or antibodies generated during the course of the studies on Saccharomyces cerevisiae conducted by the Thorner laboratory over those many years."
YeaZ system for microscopy images
YeaZ is a system for efficiently and accurately segmenting microscopy images of yeast cells. It contains a convolutional neural network, with an underlying training set of high-qual|ty segmented yeast images, as well as a graphical user interface and a web application to employ, test, and expand the system. The system contains a Python based application with graphical user interface available on GitHub, as well as standalone apps for both Windows and Mac based computers, and training sets. Additional information is available in the accompanying Nature Communications paper by Dietler et al., 2020.
YeaZ was created at École polytechnique fédérale de Lausanne (EPFL), Lausanne, Switzerland. Please contact Sahand Jamal Rahi with questions, or ideas for improvements.
In Memoriam: Angelika Amon
It was with great sadness that we learned that Angelika Amon, cell biologist and Professor of Biology at MIT and a member of the Koch Institute for Integrative Cancer Research, passed away on October 29, 2020. Angelika was widely known for her novel contributions to the field of cell biology and proliferation, with her research focusing on aneuploidy and the consequences of chromosome mis-segregation. At the beginning of Angelika's career, her work on yeast genetics led to the discovery that cyclins must break down completely before cells progress from mitosis to G1.
Her research has helped shape the current understanding of cell division, and her passion for genetics will live on through her students and colleagues who continue her work. Angelika was a treasured part of the genetics community and will be missed dearly.
The Genetics Society of America has featured a tribute piece in remembrance of Angelika Amon, written by her friend and colleague, Orna Cohen-Fix.
Gene Ontology Consortium Meeting
From October 6th-8th, PI Mike Cherry, Senior Biocuration Scientist Edith Wong, Senior Biocuration Scientist Rob Nash, Senior Biocuration Scientist Marek Skrzypek, Biocuration Scientist Suzi Aleksander, and Associate Biocuration Scientist Micheal Alexander attended the Gene Ontology Consortium meeting. Suzi presented on SGD’s GO display and GO tools, and curators learned more about how other databases utilize GO. SGD Curators also participated in meaningful discussions about improving existing GO resources while also helping with the planning of new GO projects.
We would like to thank The Gene Ontology for facilitating this successful change to a virtual conference and holding an accessible, well received event for the entire model organism community!
Happy Holidays from SGD!
We would like to take this opportunity to recognize that 2020 has brought many changes and challenges for everyone. Our thoughts go out to all those who have been impacted by the unprecedented events of this year.
We wish you and your family, friends, and lab mates the best during the upcoming holidays. Stanford University will be closed for three weeks starting on December 14 and will reopen on January 4th, 2021. Although SGD staff members will be taking time off, the website will be up and running throughout the winter break, and we will resume responding to user requests and questions in the new year.