Difference between revisions of "SGD Newsletter, Fall 2018"
(→Complex Pages Now Live) |
(→25th Anniversary of SGD) |
||
| (23 intermediate revisions by 3 users not shown) | |||
| Line 8: | Line 8: | ||
| − | SGD is proud to celebrate our 25th anniversary! | + | SGD is proud to celebrate our 25th anniversary! Initiated in 1993 for Unix workstations and later launching as a website in 1994, SGD is among the oldest biological resources on the web. The SGD website started with the basics–just check out the old CDC6 locus page! In the decades since, SGD has grown to become the premier genetics and genomics resource for ''S. cerevisiae''. |
We wish to extend a big thank you for helping us make these past decades of service to the yeast community possible. [https://twitter.com/search?q=%23apoyg #APOYG!] | We wish to extend a big thank you for helping us make these past decades of service to the yeast community possible. [https://twitter.com/search?q=%23apoyg #APOYG!] | ||
| − | <br><br><br> | + | <br><br><br><br> |
==SGD at Yeast Genetics Meeting 2018== | ==SGD at Yeast Genetics Meeting 2018== | ||
| Line 99: | Line 99: | ||
==SGD API Examples== | ==SGD API Examples== | ||
| − | [[File:sgd-api-access.jpg|left| | + | [[File:sgd-api-access-thumbnail2.jpg|border|left|190px|link=https://wiki.yeastgenome.org/images/5/53/sgd-api-access.jpg|]] |
| − | Did you know that SGD has a collection of python scripts which demonstrate how to access data from the SGD API? | + | Did you know that SGD has a collection of python scripts which demonstrate how to access data from the SGD API? |
| − | |||
| + | Click on the image shown at the left for some quick examples, or view the entire collection at our GitHub repository: https://github.com/yeastgenome/sgd_api_examples. | ||
<p> | <p> | ||
| Line 109: | Line 109: | ||
==Disease Data Now Available== | ==Disease Data Now Available== | ||
| − | |||
{| width="100%" | {| width="100%" | ||
|- style="vertical-align:middle;" | |- style="vertical-align:middle;" | ||
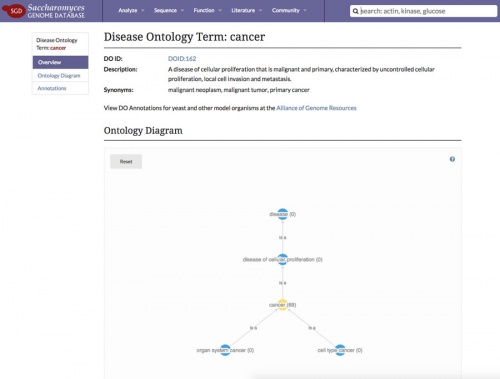
| − | |[[File:cancer_page.jpg| | + | |[[File:cancer_page.jpg|500px|link=https://www.yeastgenome.org/disease/DOID:162|thumb|center|The [https://www.yeastgenome.org/disease/DOID:162 Disease Ontology page for "cancer"]]] |
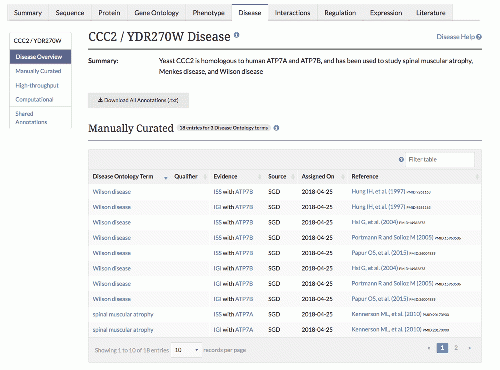
| − | |[[File:ccc2_disease.gif|thumb| | + | |[[File:ccc2_disease.gif|500px|thumb|center|caption|link=https://www.yeastgenome.org/locus/S000002678/disease|upright=1.0|The [https://www.yeastgenome.org/locus/S000002678/disease CCC2 Disease Summary page].]] |
|} | |} | ||
| − | |||
In our effort to promote the use of yeast as a catalyst for biomedical research, SGD has recently released new our Disease Pages. Each disease page corresponds to a [http://www.disease-ontology.org/ Disease Ontology] term, such as [https://www.yeastgenome.org/disease/DOID:162 cancer], and lists out all yeast genes that have a human homolog implicated in the disease. | In our effort to promote the use of yeast as a catalyst for biomedical research, SGD has recently released new our Disease Pages. Each disease page corresponds to a [http://www.disease-ontology.org/ Disease Ontology] term, such as [https://www.yeastgenome.org/disease/DOID:162 cancer], and lists out all yeast genes that have a human homolog implicated in the disease. | ||
| Line 124: | Line 122: | ||
==Complex Pages Now Live== | ==Complex Pages Now Live== | ||
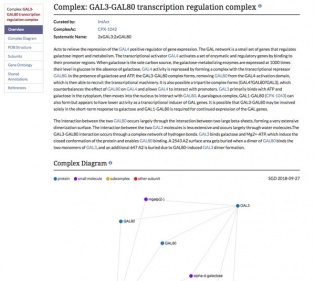
| − | [[File:complex_page.jpg|thumb| | + | [[File:complex_page.jpg|thumb|315px|right|link=https://www.yeastgenome.org/complex/CPX-1042|The [https://www.yeastgenome.org/complex/CPX-1042 GAL3-GAL80 Transcription Regulation Complex] page]] |
| − | <br> | + | <br>In addition to the new Disease data described above, SGD has also released our new Complex pages! Complex pages (example: [https://www.yeastgenome.org/complex/CPX-1042 GAL3-GAL80 complex]) provide manually curated information about the complex as well as helpful links and diagrams. Information contained within our Complex pages includes: |
| − | In addition to the new Disease data described above, SGD has also released our new Complex pages! Complex pages (example: [https://www.yeastgenome.org/complex/CPX-1042 GAL3-GAL80 complex] provide manually curated information about the complex as well as helpful links and diagrams. Information contained within our Complex pages includes: | ||
*Manually curated summaries of the complex's function and biology | *Manually curated summaries of the complex's function and biology | ||
| Line 140: | Line 137: | ||
Be sure to check out the page for your favorite complex, and let us know if you have any [mailto:sgd-helpdesk@lists.stanford.edu feedback or questions]. | Be sure to check out the page for your favorite complex, and let us know if you have any [mailto:sgd-helpdesk@lists.stanford.edu feedback or questions]. | ||
| − | + | <br> | |
| − | |||
==Website Updates== | ==Website Updates== | ||
| Line 156: | Line 152: | ||
We have also updated our [https://www.yeastgenome.org/primer3 Primer Design] tool to be based on the widely-used [http://primer3.ut.ee/ Primer3] package. It's easy to use the new [https://www.yeastgenome.org/primer3 Primer Design]. All you need to specify is the following: | We have also updated our [https://www.yeastgenome.org/primer3 Primer Design] tool to be based on the widely-used [http://primer3.ut.ee/ Primer3] package. It's easy to use the new [https://www.yeastgenome.org/primer3 Primer Design]. All you need to specify is the following: | ||
*A yeast gene name or DNA sequence of your choosing | *A yeast gene name or DNA sequence of your choosing | ||
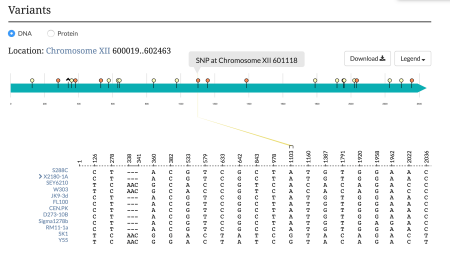
| − | *A target region to create primers for | + | *A target region to create primers for[[File:sequence-variants.png|thumb|right|upright=1.5|The [https://www.yeastgenome.org/locus/S000004218/sequence#variants ECM22 Sequence Variants section].]] |
*Optional parameters: primer length, GC content, melting temperature, and annealing. SGD has default values pre-set for these parameters, which you can change at any time. | *Optional parameters: primer length, GC content, melting temperature, and annealing. SGD has default values pre-set for these parameters, which you can change at any time. | ||
| − | |||
For more information, be sure to check out the Primer Design [https://sites.google.com/view/yeastgenome-help/analyze-help/primer-design help page]. | For more information, be sure to check out the Primer Design [https://sites.google.com/view/yeastgenome-help/analyze-help/primer-design help page]. | ||
| Line 167: | Line 162: | ||
===Upgraded GO Term Finder=== | ===Upgraded GO Term Finder=== | ||
When you've got a list of genes and want to figure out how all the genes relate to each other in terms of their molecular functions, biological processes, or cellular components, SGD's [https://www.yeastgenome.org/goTermFinder GO Term Finder] is the place to go. We've recently upgraded our GO Term Finder to be much faster and more flexible than before. Give our upgraded [https://www.yeastgenome.org/goTermFinder GO Term Finder] a spin and let us know if you have any feedback! | When you've got a list of genes and want to figure out how all the genes relate to each other in terms of their molecular functions, biological processes, or cellular components, SGD's [https://www.yeastgenome.org/goTermFinder GO Term Finder] is the place to go. We've recently upgraded our GO Term Finder to be much faster and more flexible than before. Give our upgraded [https://www.yeastgenome.org/goTermFinder GO Term Finder] a spin and let us know if you have any feedback! | ||
| − | |||
| − | |||
==Research Spotlight (Blog posts)== | ==Research Spotlight (Blog posts)== | ||
| Line 181: | Line 174: | ||
==What else have we been up to lately?== | ==What else have we been up to lately?== | ||
[[File:wellcome_KM.jpg|thumb|left|upright=.8|Kevin MacPherson at the [https://www.wellcomegenomecampus.org/ Wellcome Genome Campus]]] | [[File:wellcome_KM.jpg|thumb|left|upright=.8|Kevin MacPherson at the [https://www.wellcomegenomecampus.org/ Wellcome Genome Campus]]] | ||
| − | You may have seen SGD representatives at the following | + | <br> |
| + | You may have seen SGD representatives at the following courses: | ||
<br><br> | <br><br> | ||
| − | *Senior Biocuration Scientist Rob Nash presented a tutorial at the Yeast Genetics and Genomics course at CSHL in late July. He demonstrated the use of YeastMine and how to navigate through SGD's website. | + | *Senior Biocuration Scientist Rob Nash presented a tutorial at the [https://meetings.cshl.edu/courses.aspx?course=C-YEAS&year=19 Yeast Genetics and Genomics course] at [https://www.cshl.edu/ CSHL] in late July. He demonstrated the use of YeastMine and how to navigate through SGD's website. |
| − | *Biocuration Scientist Kevin MacPherson gave tutorials at the Fungal Pathogen Genomics course at the Wellcome Genome Campus in May. He taught students how to use tools and resources at both SGD and the Candida Genome Database. | + | *Biocuration Scientist Kevin MacPherson gave tutorials at the [https://coursesandconferences.wellcomegenomecampus.org/our-events/fungal-pathogen-genomics-2018/ Fungal Pathogen Genomics course] at the [https://www.wellcomegenomecampus.org Wellcome Genome Campus] in May. He taught students how to use tools and resources at both SGD and the [http://www.candidagenome.org/ ''Candida'' Genome Database]. |
<br><br><br> | <br><br><br> | ||
| − | |||
==Upcoming Meetings== | ==Upcoming Meetings== | ||
| Line 206: | Line 199: | ||
Ljubljana, Slovenia<br> | Ljubljana, Slovenia<br> | ||
May 22 - 24, 2019<br> | May 22 - 24, 2019<br> | ||
| + | |||
| + | <b>[http://conferences.genetics-gsa.org/tagc/2020/index The Allied Genetics Conference (TAGC 2020)]</b><br> | ||
| + | Gaylord National Resort & Convention Center, Metro Washington, DC, UCA<br> | ||
| + | April 22 – 26, 2020<br> | ||
Latest revision as of 14:17, 4 October 2018
About this newsletter:
This is the Fall 2018 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community.
Contents
25th Anniversary of SGD
SGD is proud to celebrate our 25th anniversary! Initiated in 1993 for Unix workstations and later launching as a website in 1994, SGD is among the oldest biological resources on the web. The SGD website started with the basics–just check out the old CDC6 locus page! In the decades since, SGD has grown to become the premier genetics and genomics resource for S. cerevisiae.
We wish to extend a big thank you for helping us make these past decades of service to the yeast community possible. #APOYG!
SGD at Yeast Genetics Meeting 2018
From August 22-26, 2018, the SGD team was out in full force at the Yeast Genetics Meeting in Stanford, CA. Here are some of the highlights from this awesome meeting:

Mike Cherry Receives Ira Herskowitz Award
For his outstanding contributions in the field of yeast research in the last few decades, Mike Cherry, PI of SGD, received the Ira Herskowitz Award. Congratulations Mike!
Workshop: Getting More out of SGD
At our workshop, we talked about new features, cool tools, and some of the many ways you can access data at SGD to support your research. Check out the links below to download presentations given by SGD staff:
| Presenter | Presentation Title |
| Kevin MacPherson | Tools and Resources |
| Patrick Ng | Looking at Loci with SGD |
| Edith Wong | Finding Data(files) at SGD |
| Stacia Engel | Alliance of Genome Resources |
Posters
Below is a collection of posters SGD staff presented at the Yeast Genetics Meeting. Click on any of the links below to download the poster.
| Presenter | Poster Title |
| Suzi Aleksander | The Use of the Gene Ontology to Describe Biological Function at SGD |
| Barbara Dunn | Beyond S288C: Incorporating new S. cerevisiae strain genomes and their associated not-in-S288C ORFs into SGD |
| Stacia Engel | Updated regulation curation model at the Saccharomyces Genome Database |
| Felix Gondwe | Downloading Data from SGD |
| Kalpana Karra | YeastMine: An Advanced Search and Analysis Tool for SGD Data |
| Kevin MacPherson | Saccharomyces Genome Database: Outreach and online training services |
| Rob Nash | Associating Yeast Genes with Human Disease-related Genes at SGD |
| Patrick Ng | Comparative Genomics at the Saccharomyces Genome Database |
| Ajay Shrivatsav VP | SGD Migration to AWS Architecture |
| Travis Sheppard | Getting Data: API Access to SGD and The Alliance of Genome Resources |
| Edith Wong | Forming Protein Complex Pages at the Saccharomyces Genome Database |
Fun Stuff
A big thank you to all who stopped by our booth! Visitors received all sorts of informational handouts and fun prizes, such as a SuperBud yeast toy. Many were also up to the challenge of completing our Yeast Genetics Crossword puzzle… and to the victors went the spoils of an awesome 25th anniversary SGD T-shirt!
We are always interested in answering questions and meeting the researchers and users that make up our community. We appreciate hearing your thoughts and all the suggestions you shared with us. For those of you who couldn't make it or didn't get a chance to stop by our booth, please feel free to share any comments or suggestions by contacting us!
If you would like to see tweets from the conference, search #Yeast18 on Twitter.
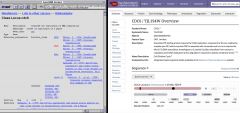
SGD API Examples
Did you know that SGD has a collection of python scripts which demonstrate how to access data from the SGD API?
Click on the image shown at the left for some quick examples, or view the entire collection at our GitHub repository: https://github.com/yeastgenome/sgd_api_examples.
Disease Data Now Available
In our effort to promote the use of yeast as a catalyst for biomedical research, SGD has recently released new our Disease Pages. Each disease page corresponds to a Disease Ontology term, such as cancer, and lists out all yeast genes that have a human homolog implicated in the disease.
Yeast genes with human homologs implicated in disease will also have a new Disease Summary tab (example: CCC2). All associated diseases are shown, along with a network diagram that relates yeast genes, their human homologs, and the diseases associated with the human homologs.
For more information, be sure to check out our Disease Ontology help page. Check out the new pages, and be sure to let us know if you have any questions or comments!
Complex Pages Now Live
In addition to the new Disease data described above, SGD has also released our new Complex pages! Complex pages (example: GAL3-GAL80 complex) provide manually curated information about the complex as well as helpful links and diagrams. Information contained within our Complex pages includes:
- Manually curated summaries of the complex's function and biology
- A list of all subunits and other complex participants
- A Complex Diagram that shows the physical interactions between each subunit
- Gene Ontology (GO) terms annotated to the complex
- Images of complex structure from the Protein Data Bank, if available
- A network diagram that shows how the complex relates to other complexes in terms of function and shared subunits
Complex pages can be accessed by running a search for the complex, or by visiting the gene pages of its subunits. For example, to visit the GAL3-GAL80 complex page, simply run a search for "GAL3-GAL80" and click on the Complexes category. Or, go to the GAL3 or GAL80 gene page and locate the Complex section.
Be sure to check out the page for your favorite complex, and let us know if you have any feedback or questions.
Website Updates
You may have noticed that a lot of SGD tools and features have been getting big updates. Here are some of the latest updates you can try out and explore:
New and Improved Gene Sequence Resources
SGD's Gene/Sequence Resources (GSR) tool is a great way to figure out which tools at SGD can help you get information on the yeast genome. Given a chromosomal region, or raw DNA/protein sequence, GSR retrieves a list of sequence analysis tools, options for accessing biological information, and table/map displays. We’ve recently updated Gene/Sequence Resources to expand its search capabilities and provide more options for downloading, analyzing, and comparing sequences from different S. cerevisiae strains. New features in Gene/Sequence Resources include:
- Options to select up to 11 different yeast strains for gene searches and sequence downloading
- Ability to search for multiple genes at a time
- Sequence alignment/variation tools such as Variant Viewer
- Convenient, automatic loading of your sequence/gene of interest into other tools at SGD
Primer Design Update
We have also updated our Primer Design tool to be based on the widely-used Primer3 package. It's easy to use the new Primer Design. All you need to specify is the following:
- A yeast gene name or DNA sequence of your choosing
- A target region to create primers for
- Optional parameters: primer length, GC content, melting temperature, and annealing. SGD has default values pre-set for these parameters, which you can change at any time.
For more information, be sure to check out the Primer Design help page.
Variant Information in Sequence Tab
On the Sequence summary tab for your favorite gene, we've added a new feature that lets you compare gene sequence variants between twelve commonly used S. cerevisiae strains. The Variants graph, derived from our Variant Viewer tool, depicts all nucleotide or amino acid changes relative to strain S288C in an easy-to-read format. To find this information for your favorite gene, go to its gene summary page and click on the "Sequence" tab. The feature will be displayed in the Variants section (example: ECM22 Sequence summary tab)
Upgraded GO Term Finder
When you've got a list of genes and want to figure out how all the genes relate to each other in terms of their molecular functions, biological processes, or cellular components, SGD's GO Term Finder is the place to go. We've recently upgraded our GO Term Finder to be much faster and more flexible than before. Give our upgraded GO Term Finder a spin and let us know if you have any feedback!
Research Spotlight (Blog posts)
In case you missed them, here are some of the most popular Research Spotlights posted on our blog lately:
- Resistance is Not Futile: How Yeasts Avoid Getting Ill from IILs Higgins and colleagues uncover ways to render yeast resistant to ionic liquids, paving the path for more efficient biofuel production.
- Mixing Mitochondria Makes Magic Wolters and coworkers show that many yeast cells recombine mitochondrial DNA after mating, for better or for worse.
- Too Much of a Good Thing Can Be Hazardous to Your Health Chakraborty and colleagues reveal that making too much of DNA mismatch repair proteins can be disastrous for your DNA and your health.
- (Almost) All Hands on Deck for Calcineurin Connolly and colleagues uncover the roles of the four mysterious "arms" of the yeast calcineurin protein, Cnb1p.
- Chaperone Wars: The Last Co-chaperone Reidy and coworkers show that Sti1p helps save the galaxy by helping a protein chaperone perform its function and interact with other proteins.
What else have we been up to lately?

You may have seen SGD representatives at the following courses:
- Senior Biocuration Scientist Rob Nash presented a tutorial at the Yeast Genetics and Genomics course at CSHL in late July. He demonstrated the use of YeastMine and how to navigate through SGD's website.
- Biocuration Scientist Kevin MacPherson gave tutorials at the Fungal Pathogen Genomics course at the Wellcome Genome Campus in May. He taught students how to use tools and resources at both SGD and the Candida Genome Database.
Upcoming Meetings
EMBO Workshop on Experimental Approaches to Evolution and Ecology Using Yeast and Other Model Systems
Heidelberg, Germany
October 17 - 20, 2018
7th International Yeast 2.0 and Synthetic Genomes Conference
Sydney, NSW, Australia
November 26 - 28, 2018
30th Fungal Genetics Conference
Asilomar Conference Grounds, Pacific Grove, CA, USA
March 12 - 17, 2019
14th Yeast Lipid Conference - YLC 2019
Ljubljana, Slovenia
May 22 - 24, 2019
The Allied Genetics Conference (TAGC 2020)
Gaylord National Resort & Convention Center, Metro Washington, DC, UCA
April 22 – 26, 2020