Difference between revisions of "YJL133C-A"
SGDwikiBot (talk | contribs) (Automated import of articles) |
|||
| (48 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
{|{{Prettytable}} align = 'right' width = '200px' | {|{{Prettytable}} align = 'right' width = '200px' | ||
|- | |- | ||
| − | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http:// | + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Systematic name''' || [http://www.yeastgenome.org/cgi-bin/locus.pl?dbid=S000028805 YJL133C-A] |
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Gene name''' ||'' '' | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Coordinates''' | ||
| − | |nowrap| Chr X: | + | |nowrap| Chr X:159847..159623 |
| + | |- | ||
| + | |valign="top" nowrap bgcolor="{{SGDblue}}"| '''Primary SGDID''' || S000028805 | ||
|} | |} | ||
<br> | <br> | ||
| − | '''Description of | + | '''Description of YJL133C-A:''' Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies<ref name='S000117178'>Reinders J, et al. (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5(7):1543-54 |
{{SGDpaper|S000117178}} PMID 16823961</ref> | {{SGDpaper|S000117178}} PMID 16823961</ref> | ||
<br> | <br> | ||
| Line 27: | Line 29: | ||
==Community Commentary== | ==Community Commentary== | ||
{{CommentaryHelp}} | {{CommentaryHelp}} | ||
| + | |||
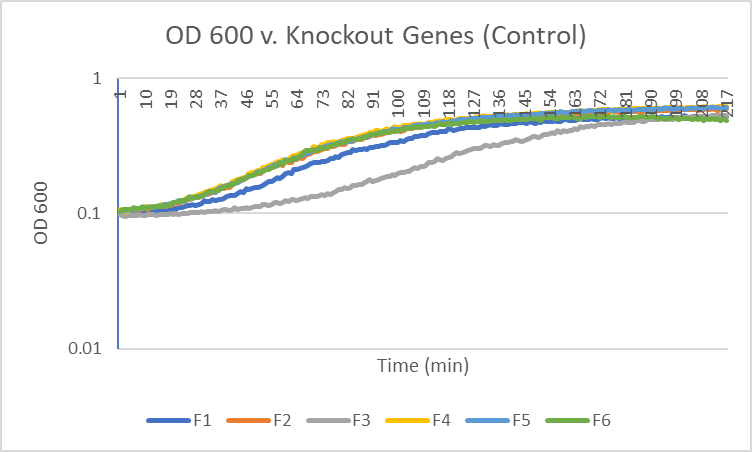
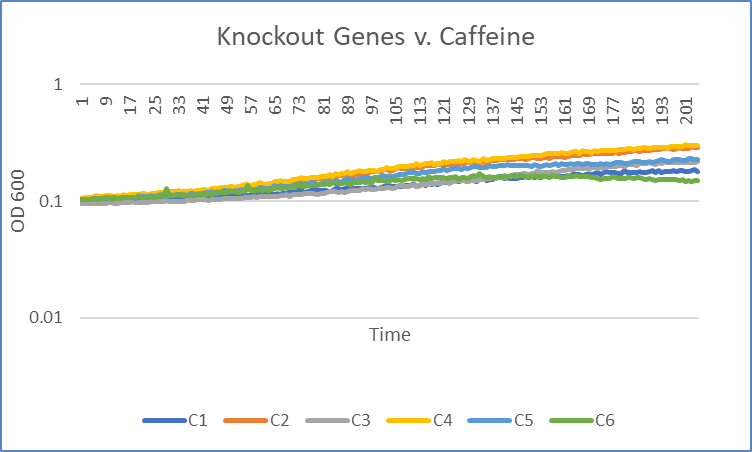
| + | ===[[UW-Stout/Caffeine_FA22|Caffeine]]=== | ||
| + | |||
| + | [[Image:F_graph.png|100pxs|Control]] | ||
| + | |||
| + | [[Image:Knockout_v._caffeine_trial1_Viv.png|100pxs|Trial 1]] | ||
| + | |||
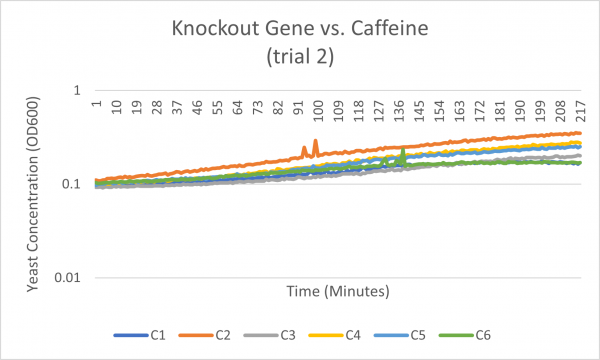
| + | [[Image:Knockout_gene_v._Caffeine_trial_2.png|600px|Trial 2]] | ||
| + | |||
| + | ====Interpretation==== | ||
| + | |||
| + | The average doubling time for the YJL133C-A (C2) gene in standard conditions was 89.5 minutes. Under caffeine stress, the average doubling time was 134 minutes. That is a 49.7% increase in doubling time which means that cell growth was negatively affected (slowed down) by this type of stress. | ||
| + | |||
| + | ===[[UW-Stout/Ethanol_FA22|Ethanol]]=== | ||
| + | |||
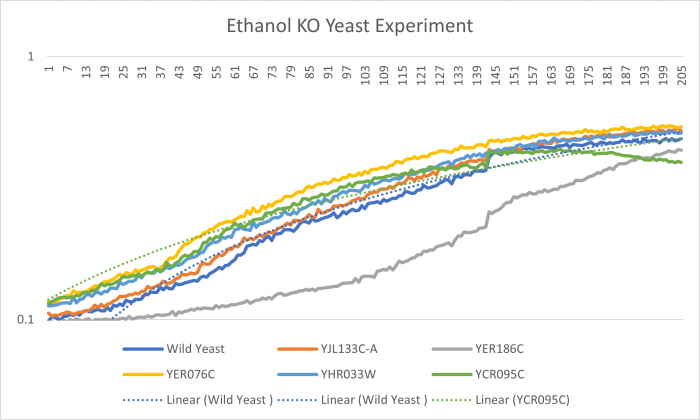
| + | [[Image:Best Ethanol KO graph.png|Graph|center|700px|YJL133C-A]] | ||
| + | |||
| + | YJL133C-A's line lies in the middle of the overall growth curve. The doubling time is 67.78. These cells went not affected but the Ethanol. | ||
| + | |||
| + | ===[[UW-Stout/Ultraviolet_Light_FA22|UV Light]]=== | ||
| + | |||
| + | [[Image:BY4735_FA22_UV_NegControl.png|left|thumb|300px|Wild type yeast prepared according to sample prep. No UV exposure. 72 colonies.]][[Image:BY4735_FA22_UV_PosControl.png|center|thumb|300px|Wild type yeast prepared according to sample prep and UV exposure protocol. 11 colonies.]] | ||
| + | |||
| + | [[Image:YJL133C-A FA22 UV NegControl.png|left|thumb|300px|YJL133C-A knock-out yeast prepared according to sample prep. No UV exposure. 68 colonies.]] | ||
| + | [[Image:YJL133C-A FA22 UV Trial1.png|center|thumb|300px|YJL133C-A knock-out yeast prepared according to sample prep and UV exposure protocol. Trial 1. 13 colonies.]] | ||
| + | [[Image:YJL133C-A FA22 UV Trial2.png|right|thumb|300px|YJL133C-A knockout yeast prepared according to sample prep and UV exposure protocol. Trial 2. 7 colonies.]] | ||
| + | |||
| + | |||
| + | The plate with the modified yeast that was not exposed to UV had approximately 5% fewer colonies than the wild-type plate that was not exposed to UV. This shows that the knock-out strain of yeast is expected to be less viable compared to wild-type yeast in the absence of stressors. | ||
| + | The two trial plates were pretty similar in colony count. The average of the two was 10 colonies. | ||
| + | The plate with wild-type yeast that was exposed to UV had 11 colonies. | ||
| + | There was no significant difference in colony count between the modified yeast and wild-type yeast that was exposed to UV. | ||
| + | |||
| + | |||
| + | As there was no significant difference between the wild-type yeast exposed to UV and the trial strains, we can conclude that the gene is not used in the defense against UV damage nor the repair of UV damage in yeast. | ||
| + | |||
| + | ===Hydrogen Peroxide=== | ||
| + | [[Image:1.jpg|center|thumb|600px|The x-axis is time in minutes and the y-axis is OD600]] | ||
| + | |||
| + | This is the link to the protocol page: [[UW-Stout/Hydrogen Peroxide FA22]]. | ||
| + | |||
| + | The doubling time for YJL133C-A under the stress of hydrogen peroxide is 306 minutes whereas without stress it was only 206 minutes. As shown in the chart above, the optical density of the YJL133C-A is very similar to that of the wild type yeast. The BY4375 under stress had a 598 minute doubling time and with no stress there was a 229 minute doubling time. Even though the stress placed on the YJL133C-A yeast, resulted in an increased doubling time, this was consistent with the wild type yeast. | ||
| + | |||
| + | |||
| + | <!-- PLEASE ADD Community Commentary ABOVE THIS MESSAGE. See below for an example of community annotation --> | ||
| + | <!-- | ||
| + | Specifically higher expression in carbon limited chemostat cultures versus carbon excess. | ||
| + | <ref>Boer VM, et al. (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. | ||
| + | J Biol Chem 278(5):3265-74</ref> | ||
| + | --> | ||
| + | |||
<protect> | <protect> | ||
| + | |||
==References== | ==References== | ||
<!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | <!-- REFERENCES ARE AUTOMATICALLY GENERATED. PLEASE DON'T EDIT THIS SECTION--> | ||
{{RefHelp}} | {{RefHelp}} | ||
</protect> | </protect> | ||
| + | |||
| + | ==UW Stout/FA 2022 Glycerol Media== | ||
| + | ===Introduction=== | ||
| + | Following the [[UW-Stout/Glycerol FA22]] protocol, 5 knockout strains of yeast cell were tortured within a glycerol media solution. | ||
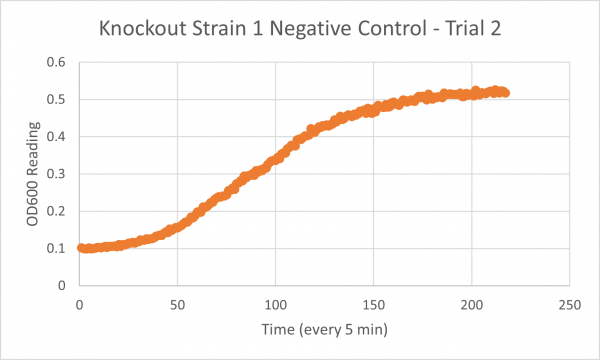
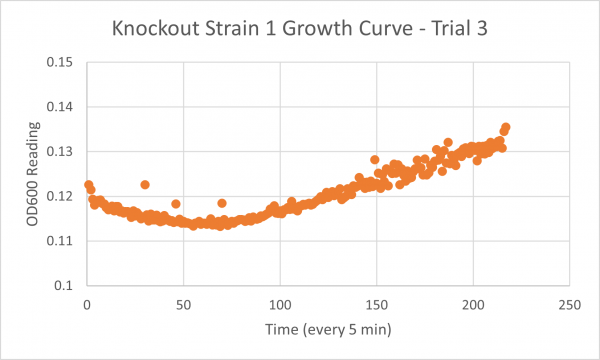
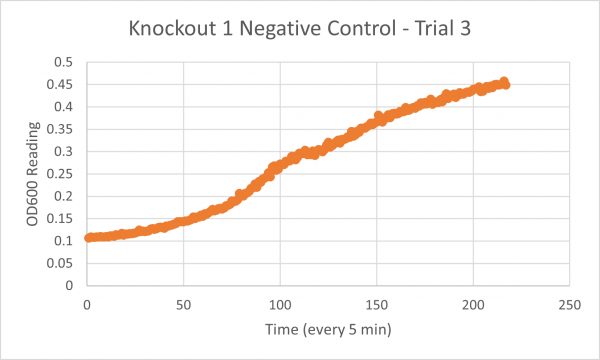
| + | ===Results=== | ||
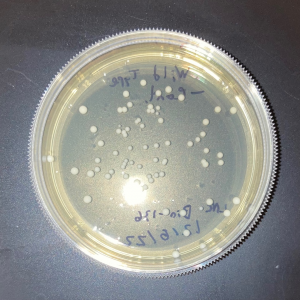
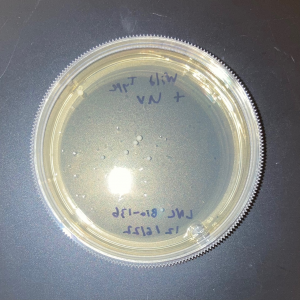
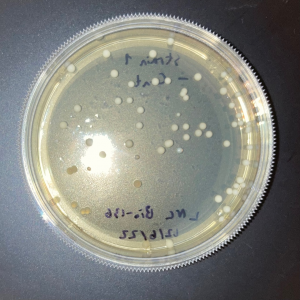
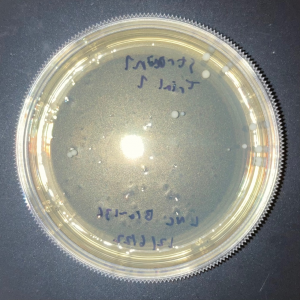
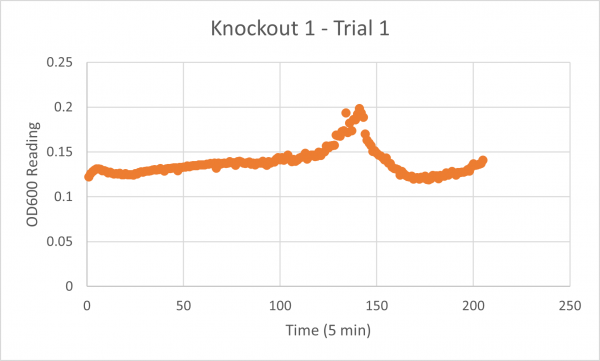
| + | [[Image:Glycerol_Knockout_1_Trial_1.png|600px]] | ||
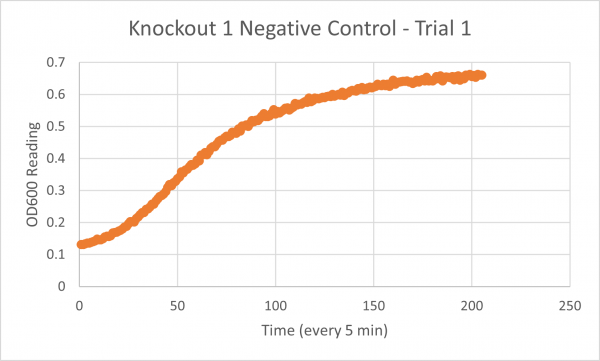
| + | [[Image:Negative Control KO1 Trial 1.png|600px]] | ||
| + | [[Image:Glycerol_Knockout_1_Trial_2.png|600px]] | ||
| + | [[Image:Negative_Control_KO1_T2.png|600px]] | ||
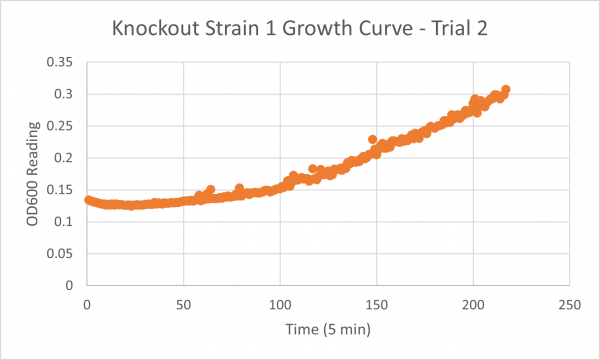
| + | [[Image:Glycerol Growth_Curve_KO1_T3.png|600px]] | ||
| + | [[Image:Negative_Control_KO1_Trial_3.png|600px]] | ||
| + | |||
| + | The following doubling times were calculated: | ||
| + | * Glycerol Trial 1: NA | ||
| + | * Negative Control Trial 1: 163 min | ||
| + | * Glycerol Trial 2: 565 min | ||
| + | * Negative Control Trial 2: 211 min | ||
| + | * Glycerol Trial 3: NA | ||
| + | * Negative Control Trial 3: 236 min | ||
| + | |||
| + | ===Analysis/Conclusion=== | ||
| + | Trial 1 and trial 3 of the glycerol torture experiment performed on the knockout on the YJL133C-A in yeast did not look incredibly promising- it is hard to rely on these results and draw any conclusions. The main pattern that seems to be apparent is that the yeast is not reproducing at a consistent rate when grown in glycerol without this gene functioning. A number of different factors could have gone wrong for these results to turn out this way, so it is hard to tell what happened! Disregarding trial 1 and trial 3, the data in trial 3 seemed promising! It is clear that the yeast was growing at a much slower rate, with less than half of a doubling time than what appeared in the negative control. I would definitely say that growing yeast in a glycerol media without YJL133C-A functioning puts a significant amount of stress on the organism. | ||
| + | |||
| + | ==UW Stout/FA 2022 Bud Scars== | ||
| + | Cells that had the gene YJL133C-A had an average of 1.06 scars per cell. Out of 100 cells counted, 106 bud scars were counted. The wild yeast BY4735 had an average of 2.46 bud scars per cell (100 cells counted, 246 bud scars). This indicates that the knocked-out gene significantly slowed down the reproduction rate of these cells. | ||
| + | [[File:Yeast_1(2)_2_67.png]] | ||
| + | ''picture does not include all cells that were counted and added for visual of bud scars'' | ||
Latest revision as of 17:48, 20 December 2022
Share your knowledge...Edit this entry! <protect>
| Systematic name | YJL133C-A |
| Gene name | |
| Aliases | |
| Feature type | ORF, Uncharacterized |
| Coordinates | Chr X:159847..159623 |
| Primary SGDID | S000028805 |
Description of YJL133C-A: Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies[1]
</protect>
Contents
Community Commentary
About Community Commentary. Please share your knowledge!
Caffeine
Interpretation
The average doubling time for the YJL133C-A (C2) gene in standard conditions was 89.5 minutes. Under caffeine stress, the average doubling time was 134 minutes. That is a 49.7% increase in doubling time which means that cell growth was negatively affected (slowed down) by this type of stress.
Ethanol
YJL133C-A's line lies in the middle of the overall growth curve. The doubling time is 67.78. These cells went not affected but the Ethanol.
UV Light
The plate with the modified yeast that was not exposed to UV had approximately 5% fewer colonies than the wild-type plate that was not exposed to UV. This shows that the knock-out strain of yeast is expected to be less viable compared to wild-type yeast in the absence of stressors.
The two trial plates were pretty similar in colony count. The average of the two was 10 colonies.
The plate with wild-type yeast that was exposed to UV had 11 colonies.
There was no significant difference in colony count between the modified yeast and wild-type yeast that was exposed to UV.
As there was no significant difference between the wild-type yeast exposed to UV and the trial strains, we can conclude that the gene is not used in the defense against UV damage nor the repair of UV damage in yeast.
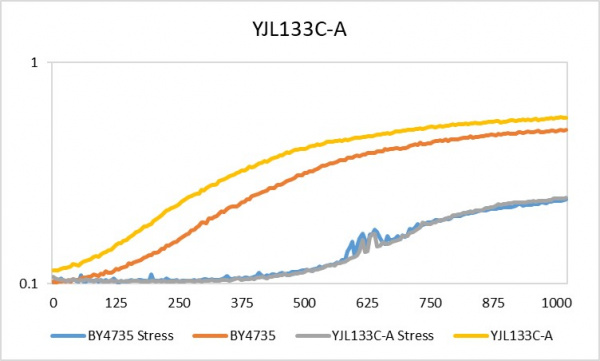
Hydrogen Peroxide
This is the link to the protocol page: UW-Stout/Hydrogen Peroxide FA22.
The doubling time for YJL133C-A under the stress of hydrogen peroxide is 306 minutes whereas without stress it was only 206 minutes. As shown in the chart above, the optical density of the YJL133C-A is very similar to that of the wild type yeast. The BY4375 under stress had a 598 minute doubling time and with no stress there was a 229 minute doubling time. Even though the stress placed on the YJL133C-A yeast, resulted in an increased doubling time, this was consistent with the wild type yeast.
<protect>
References
See Help:References on how to add references
See Help:Categories on how to add the wiki page for this gene to a Category </protect>
UW Stout/FA 2022 Glycerol Media
Introduction
Following the UW-Stout/Glycerol FA22 protocol, 5 knockout strains of yeast cell were tortured within a glycerol media solution.
Results
The following doubling times were calculated:
- Glycerol Trial 1: NA
- Negative Control Trial 1: 163 min
- Glycerol Trial 2: 565 min
- Negative Control Trial 2: 211 min
- Glycerol Trial 3: NA
- Negative Control Trial 3: 236 min
Analysis/Conclusion
Trial 1 and trial 3 of the glycerol torture experiment performed on the knockout on the YJL133C-A in yeast did not look incredibly promising- it is hard to rely on these results and draw any conclusions. The main pattern that seems to be apparent is that the yeast is not reproducing at a consistent rate when grown in glycerol without this gene functioning. A number of different factors could have gone wrong for these results to turn out this way, so it is hard to tell what happened! Disregarding trial 1 and trial 3, the data in trial 3 seemed promising! It is clear that the yeast was growing at a much slower rate, with less than half of a doubling time than what appeared in the negative control. I would definitely say that growing yeast in a glycerol media without YJL133C-A functioning puts a significant amount of stress on the organism.
UW Stout/FA 2022 Bud Scars
Cells that had the gene YJL133C-A had an average of 1.06 scars per cell. Out of 100 cells counted, 106 bud scars were counted. The wild yeast BY4735 had an average of 2.46 bud scars per cell (100 cells counted, 246 bud scars). This indicates that the knocked-out gene significantly slowed down the reproduction rate of these cells.
 picture does not include all cells that were counted and added for visual of bud scars
picture does not include all cells that were counted and added for visual of bud scars