Difference between revisions of "YeaZ"
(Created page with "https://www.epfl.ch/labs/lpbs/data-and-software/|YeaZ] is a system for efficiently and accurately segmenting microscopy images of yeast cells. It contains a convolutional neur...") |
|||
| Line 1: | Line 1: | ||
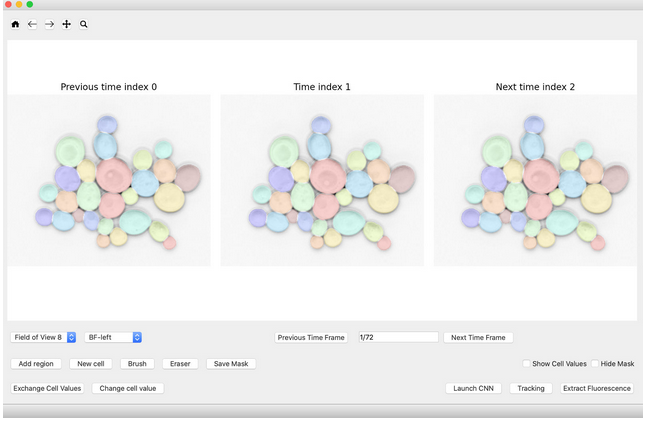
https://www.epfl.ch/labs/lpbs/data-and-software/|YeaZ] is a system for efficiently and accurately segmenting microscopy images of yeast cells. It contains a convolutional neural network, with an underlying training set of high-quality segmented yeast images, as well as a graphical user interface and a web application to employ, test, and expand the system. | https://www.epfl.ch/labs/lpbs/data-and-software/|YeaZ] is a system for efficiently and accurately segmenting microscopy images of yeast cells. It contains a convolutional neural network, with an underlying training set of high-quality segmented yeast images, as well as a graphical user interface and a web application to employ, test, and expand the system. | ||
| − | [[sgdwiki-yeaz-screenshot.png]] | + | [[Image:sgdwiki-yeaz-screenshot.png]] |
The system contains a Python based application with graphical user interface available on GitHub (https://github.com/lpbsscientist/YeaZ-GUI), as well as standalone apps for both Windows and Mac based computers, as well as training sets. Additional information is available in the accompanying Nature Communications paper by [https://www.nature.com/articles/s41467-020-19557-4|Dietler et al., 2020](https://www.nature.com/articles/s41467-020-19557-4). | The system contains a Python based application with graphical user interface available on GitHub (https://github.com/lpbsscientist/YeaZ-GUI), as well as standalone apps for both Windows and Mac based computers, as well as training sets. Additional information is available in the accompanying Nature Communications paper by [https://www.nature.com/articles/s41467-020-19557-4|Dietler et al., 2020](https://www.nature.com/articles/s41467-020-19557-4). | ||
Revision as of 10:35, 13 November 2020
https://www.epfl.ch/labs/lpbs/data-and-software/%7CYeaZ] is a system for efficiently and accurately segmenting microscopy images of yeast cells. It contains a convolutional neural network, with an underlying training set of high-quality segmented yeast images, as well as a graphical user interface and a web application to employ, test, and expand the system.
The system contains a Python based application with graphical user interface available on GitHub (https://github.com/lpbsscientist/YeaZ-GUI), as well as standalone apps for both Windows and Mac based computers, as well as training sets. Additional information is available in the accompanying Nature Communications paper by et al., 2020(https://www.nature.com/articles/s41467-020-19557-4).
YeaZ was created at École polytechnique fédérale de Lausanne (EPFL), Lausanne, Switzerland
Please contact Sahand Jamal Rahi with questions, or ideas for improvements.