Difference between revisions of "What are yeast?"
(→General Information) |
|||
| (93 intermediate revisions by 7 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | ====General | + | ====General Introduction==== |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | Yeast are | + | Most of us know yeast is a very helpful organism, especially with respect to baking, wine making, and brewing. However, what are yeast and why are they the focus of so much research? |
| − | warm-blooded animals, where they may live symbiotically or as parasites. | + | |
| − | is typically | + | [[File:Fungi.jpeg|thumb|left|225px|'''Introduction to Fungi''' Credit: ''Kandis Elliot, Mo Fayyaz, UW-Madison'']] |
| − | + | ||
| + | =====Yeast are Fungi===== | ||
| + | |||
| + | Yeast are single-celled microorganisms that are classified, along with molds and mushrooms, as members of the Kingdom Fungi. Yeasts are evolutionarily diverse and are therefore classified into two separate phyla, [http://en.wikipedia.org/wiki/Ascomycota ''Ascomycota''] or sac fungi and [http://en.wikipedia.org/wiki/Basidiomycota ''Basidiomycota''] or higher fungi, that together form the subkingdom [http://en.wikipedia.org/wiki/Dikarya ''Dikarya'']. Budding yeast, also referred to as “true yeasts”, are members of the phylum ''Ascomycota'' and the order ''Saccharomycetales''. Such classifications are based on characteristics of the cell, ascospore, and colony, as well as cellular physiology. | ||
| + | |||
| + | =====Yeast are Single-celled, but with Cellular Organization Similar to Higher Organisms===== | ||
| + | [[File:500px-Celltypes.jpg|thumb|right|275px|'''Yeast are single-celled organisms classified as eukaryotes due the presence of a nucleus that harbors their genetic information. ''' Credit: ''Wikicommons'']] | ||
| + | Although yeast are single-celled organisms, they possess a cellular organization similar to that of higher organisms, including humans. Specifically, their genetic content is contained within a nucleus. This classifies them as eukaryotic organisms, unlike their single-celled counterparts, bacteria, which do not have a nucleus and are considered prokaryotes. | ||
| + | |||
| + | =====Natural Habitats===== | ||
| + | |||
| + | Yeast are widely dispersed in nature with a wide variety of habitats. They are commonly found on plant leaves, flowers, and fruits, as well as in soil. Yeast are also found on the surface of the skin and in the intestinal tracts of warm-blooded animals, where they may live symbiotically or as parasites. The common "yeast infection" is typically caused by ''Candida albicans''. In addition to being the causative agent in vaginal yeast infections, ''Candida'' is also the cause of diaper rash and thrush of the mouth and throat. | ||
| + | |||
| + | ====Why Study Yeast?==== | ||
| + | |||
| + | Imagine an organism that grows quickly in a flask and whose DNA can be easily manipulated, but also provides insight into basic human biological processes, including disease. Yeast fits that description and is the focus of study for researchers all over the world, resulting in more than 50,000 published scientific articles describing yeast research! | ||
| + | |||
| + | [[File:Grapes.jpg|thumb|left|160px|'''Yeast are commonly found on grapes.''' Credit: ''Wikicommons'']] | ||
| + | |||
| + | |||
| + | What specific characteristics of yeast make it a “model organism” for study and the focus of so much research? Yeast are single-celled (unicellular) organisms, making them simple to study, but possess a cellular organization similar to that found in higher, multi-cellular organisms such as humans – that is, they possess a nucleus and are therefore eukaryotes, as described above. Most importantly, the similarity in cellular organization between yeast and higher eukaryotes translates to similarities in their fundamental cellular processes so discoveries in yeast frequently provide direct or indirect clues as to how biological processes work in humans. | ||
| + | |||
| + | [[File:DNA-molecule.jpg|thumb|right|150px|'''Ball and stick model of DNA.''' Credit: ''Wikicommons'']] | ||
| + | |||
| + | |||
| + | Another important characteristic of yeast essential to their role as “model organisms” is the fact they are relatively easy to work with. Yeast replicate quickly and are easy to manipulate genetically. The doubling time for yeast (the time required for a cell to duplicate and divide itself) is about 90 minutes. In contrast, human cells growing in culture need about 24 hours to double. There are also well defined genetic methods for yeast that allow researchers to easily isolate mutants, cross them with other mutants or into other genetic backgrounds, and map the locations of genes. In fact, genetic maps constructed based on the genetic distance between genes gave researchers their first view of the genome and its organization, and were the culmination of genetic studies that date back to the first half of the twentieth century. | ||
| + | |||
| + | |||
| + | An accelerated pace of discovery was made possible after the baker's yeast (''S. cerevisiae'') genome, representing its complete set of genetic material, became the first eukaryotic genome to be sequenced back in 1996. It is smaller and more compact than the human genome (12 million base pairs and ~6,000 genes, compared with 3 billion base pairs and ~20-25,000 protein coding genes). Yet, comparisons of the genomes indicate that ~31% of yeast genes are very similar to human genes and 20% of human disease genes have counterparts in yeast. In addition, yeast cells can exist either as haploids (one set of chromosomes) or diploids (two sets of chromosomes). Since haploids have only one copy of each gene and efficient breaking and rejoining of DNA strands (recombination), it is very easy to delete a specific gene in a haploid and observe the effects on the cell, or the “phenotype” of the deletion mutant. Diploid cells, on the other hand, make it possible to study essential genes (those required for growth and viability) by deleting one copy of the gene and making subtle changes in the other copy. Finally, with the information from the genome sequence an extensive toolkit of molecular reagents and genome-wide collections have been constructed, providing researchers with powerful means to study biological problems. If a yeast gene is known to be similar in DNA sequence to a human gene, studies in yeast can provide powerful clues as to the role of the related gene(s) in humans. Thus, the relative simplicity of studying cellular functions in yeast combined with its relevance to higher organisms makes it a very powerful “model organism” for study. | ||
| + | |||
| + | ====Yeast Life and Cell Cycles==== | ||
| + | [[File:Yeast_life_cycle.png|thumb|left|160px|'''Budding yeast life cycle.''' Credit: ''Wikicommons'']] | ||
| + | |||
| + | Yeast typically grow asexually by budding. A small bud which will become the daughter cell is formed on the parent (mother) cell, and enlarges with continued grow. As the daughter cell grows, the mother cell duplicates and then segregates its DNA. The nucleus divides and migrates into the daughter cell. Once the bud contains a nucleus and reaches a certain size it separates from the mother cell. The series of events that occur in a cell and lead to duplication and division are referred to as the cell cycle. The cell cycle consists of four distinct phases (G1, S, G2 and M) and is regulated similar to that of the cell cycle in larger eukaryotes. As long as adequate nutrients such as sugar, nitrogen and phosphate are present yeast cells will continue to divide asexually. | ||
| + | |||
| + | [[File:shmooing yeast.jpg|thumb|right|150px|'''Shmooing yeast cell.''' Credit: ''Wikicommons'']] | ||
| + | |||
| + | |||
| + | Yeast cells can also reproduce sexually. Yeast cells exist as one of two different mating types, '''a''' cells and '''alpha''' cells. When cells of opposite mating types are mixed together in the lab or randomly come into contact in nature they can mate (conjugate). Before joining the cells change shape in a process called shmooing. The term [http://en.wikipedia.org/wiki/Shmoo 'shmoo'] was coined based on its similarity in shape to that of a fictional cartoon character of the same name created back in the late 40's by Al Capp, appearing first in his comic strip L'il Abner. During conjugation the shmooing haploid cells first fuse and then their nuclei fuse, resulting in the formation of a diploid cell with two copies of each chromosome. Once formed, diploid cells can reproduce asexually by budding, similar to haploids. However, when diploid cells are starved of nutrients they undergo sporulation. During sporulation diploid cells undergo meiosis, a special form of cell division that reduces the number of chromosomes from two copies back to one copy. After meiosis the haploid nuclei produced in meiosis are packaged into four spores that contain modified cell walls, resulting in structures that are very resistant to environmental stress. These spores can survive long periods of time until conditions become more favorable, such as in the presence of improved nutrients, whereupon they are able to germinate and reproduce asexually. These different states, budding, conjugation and sporulation together make up the yeast life cycle. | ||
| + | |||
| + | [[File:CO2_bubbles.jpg|thumb|left|150px|'''CO2 bubbles produced during fermentation.''' Credit: ''Wikicommons'']] | ||
| + | |||
| + | ====Yeast growth and metabolism==== | ||
| + | When yeast cells are grown in rich carbon sources such as glucose they prefer to grow by fermentation. During fermentation glucose is converted into carbon dioxide and ethanol. Generally, fermentation occurs in the absence of oxygen, and is therefore anaerobic by nature. Even in the presence of oxygen yeast cells prefer to grow fermentatively and this is referred to as the Crabtree Effect after the biologist who discovered this preference. This form of growth is exploited in the making of bread, beer, wine and other alcoholic beverages. Although budding yeast cells prefer to grow by fermentation, when nutrients are limiting they are also able to grow by cellular respiration. During respiration cells convert glucose into carbon dioxide and water, consuming oxygen in the process, and resulting in the production of much larger amounts of energy in the form of ATP. | ||
| + | |||
| + | ====Historical Discoveries==== | ||
| + | |||
| + | [[File:Iron_age_Celtic_Brewery.jpg|thumb|right|150px|'''Egyptian wooden model of beer making in ancient Egypt.''' Credit: ''Wikicommons'']] | ||
| + | |||
| + | Yeast has been used as an industrial microorganism for 1000’s of years. The ancient Egyptians used yeast fermentation to leaven bread. There is evidence of grinding stones, baking chambers and drawings of 4000-year-old bakeries. Archaeological digs have uncovered evidence in the form of jars containing the remains of wine that is 7,000 years old. | ||
| + | |||
| + | |||
| + | Yeast were first visualized in 1680 by Antoni van Leeuwenhoek using high quality lenses. However, he thought that these globules were starchy particles of the grain used to make wort, the liquid extract used in brewing, rather than fermenting yeast cells. In 1789, Antoine Lavoisier, a French chemist, contributed to our understanding of the basic chemical reactions needed to produce alcohol from sugarcane. By estimating the proportion of starting materials and products (ethanol and carbon dioxide) after adding yeast paste he concluded that two chemical pathways were used with two thirds of the sugar reduced to alcohol and one third to form carbon dioxide. However, at the time it was thought that yeast were merely there to initiate the reaction rather than being required throughout the process. | ||
| + | |||
| + | [[File:Ascus.gif|thumb|left|150px|'''Ascus of ''S. cerevisiae'' containing a tetrad of four spores.''' Credit: ''Wikicommons'']] | ||
| + | |||
| + | |||
| + | In 1815, Joseph-Louis Gay-Lussac, a French chemist, developed methods to maintain grape juice in an unfermented state and discovered that the introduction of ‘ferment’ (which contains yeast) was required to convert unfermented wort, demonstrating the importance of yeast for alcoholic fermentation. In 1835, Charles Cagniard de la Tour used a more powerful microscope to show that yeast were single celled and multiplied by budding. In the 1850s Louis Pasteur discovered that fermented beverages resulted from the conversion of glucose to ethanol by yeast and defined fermentation as "respiration without air". Near the end of the 1800s Eduard Buchner used cell-free extracts obtained by grinding yeast cells to detect zymase, the collection of enzymes that promote or catalyze fermentation and for this he was awarded the Nobel Prize in 1907. | ||
| + | |||
| + | |||
| + | Much of the pioneering work on yeast genetics was carried out by Øjvind Winge. He discovered that yeast alternate between haploid and diploid states and that yeast are heterothallic, as two strains are required to convert haploids to diploids (conjugation). He and his colleague Otto Laustsen devised techniques to micromanipulate yeast so they could be investigated genetically. With this technique, known as "tetrad analysis", a fine needle and a microscope are used to isolate a structure known as an ascus, which contains the four spore products or tetrad resulting from sporulation of a diploid. Once the ascus is isolated, the spores in the tetrad are teased apart and allowed to grow into colonies for genetic analysis. This pioneering work earned him the title ‘The Father of Yeast Genetics’. Some of this work was further clarified by Carl Lindegren, who elucidated the mating-type system in budding yeast, demonstrating the existence of Mat '''a''' and Mat '''alpha''' cells, devised methods to carry our mass matings between cells of these mating types and used this knowledge to study the genetics of sugar utilization. | ||
| + | |||
| + | |||
| + | Since that time many other researchers have carried out groundbreaking research using budding yeast. Some of these researchers have been awarded the Nobel Prize for significant discoveries made during these studies, including: Dr. Leland Hartwell (2001) for the discovery of genes that regulate the cell cycle (co-winner with Paul Nurse and Tim Hunt); Roger Kornberg (2006) for his studies on the first step of gene expression, the means by which a genes DNA sequence is copied into messenger RNA (mRNA); Drs. Elizabeth Blackburn, Carol Greider and Jack Szostak (2009) for discovering and elucidating the genes and means by which cells protect chromosome ends or telomeres from being degraded; and to Drs. Randy Schekman, James Rothman and Thomas Südhof (2013) for research on the machinery that regulates vesicular traffic. Most recently, Dr. Yoshinori Ohsumi was awarded the prize for his work on autophagy, which began with studies in yeast. | ||
====Commercial Applications==== | ====Commercial Applications==== | ||
| − | '' | + | [[File:Beer_and_bread.jpg|thumb|right|175px|'''Yeast is used to make beer and bread.''' Credit: ''Wikicommons'']] |
| + | |||
| + | Yeast has long been considered to be the organism of choice for the production of alcoholic beverages, bread, and a large variety of industrial products. This is based on the ease with which the metabolism of yeast can be manipulated using genetic techniques, the speed with which it can be grown to high cell yields (biomass), the ease with which this biomass can be separated from products and the knowledge that it is generally recognized as safe (GRAS). | ||
| + | |||
| + | |||
| + | The budding yeast ''S. cerevisiae'' and other yeast species have long been used to ferment the sugars of rice, wheat, barley, and corn to produce alcoholic beverages such as beer and wine. There are two major types of brewing yeast, top-fermenting ale yeast and bottom-fermenting lager yeast. Top-fermenting yeast such as ''S. cerevisiae'' rise to the surface during fermentation and are used to brew ales, porters, stouts and wheat beers. In contrast, ''S. pastorianus'', (formerly known as ''S. carlsbergensis'') is a bottom-fermenting yeast used to make lager beer. Lager yeasts grow best at lower temperatures. As a result they grow more slowly, produce less surface foam, and therefore typically settle to the bottom of the fermenter. Pilsners, Märzen, Bocks, and American malt liquors are all styles of lager beer. In modern brewing many of the original top fermenting strains have been modified to become bottom fermenters. | ||
| + | |||
| + | |||
| + | Yeast produce wine by fermenting sugars from grape juice (must) into ethanol. Although wine fermentation can be initiated by naturally occurring yeast present in the vineyards, many wineries choose to add a pure yeast culture to dominate and control the fermentation. The bubbles in champagne and sparkling wines are produced by a secondary fermentation, typically in the bottle, which traps the carbon dioxide. Carbon dioxide produced in wine production is released as a by-product. One yeast cell can ferment approximately its own weight in glucose per hour. Under optimal conditions ''S. cerevisiae'' can produce up to 18 percent, by volume of ethanol with 15 - 16% being the norm. The sulfur dioxide present in commercially produced wine is added just after the grapes are crushed to kill the naturally present bacteria, mold, and yeast. | ||
| + | |||
| + | [[File:Nutritional_yeast.jpg|thumb|left|175px|'''Yeast has a nutty, cheesy flavor making it an ideal cheese substitute.''' Credit: ''Wikicommons'']] | ||
| + | |||
| + | |||
| + | ''Saccharomyces cerevisiae'' or baker’s yeast has long been used as a leavening agent in baking. Baker’s yeast ferment sugars present in dough, producing carbon dioxide and ethanol. The carbon dioxide becomes trapped in small bubbles in the dough, which causes the dough to rise. Sourdough bread is an exception, as it is not produced using baker's yeast, but is instead made with a combination of wild yeast and bacteria. The yeast ''Candida milleri'' is used to strengthen the gluten, and an acid-generating bacteria “Lactobacillus sanfranciscensis”, is used to ferment the maltose. | ||
| + | |||
| + | |||
| + | In addition to these traditional uses yeast has also been used for many other commercial applications. Vegans often use yeast as a cheese substitute and it is often used as a topping for products such as popcorn. It is being used in the petrochemical industry where it has been engineered to produce biofuels such as ethanol, and farnesene, a diesel and jet fuel precursor. It is also used in the production of lubricants and detergents. Yeast is used in the food industry for the production of food additives including colorants, antioxidants, and flavor enhancers. It is the often used in the production of pharmaceuticals including antiparasitics, anticancer compounds, biopharmaceuticals such as insulin, vaccines, and nutraceuticals. Yeast is commonly used in the production of industrial enzymes and chemicals. In the field of environmental bioremediation strains have even been exploited for the removal of metal from mining waste. | ||
| + | |||
| + | ====Application to Human Disease and Research==== | ||
| + | |||
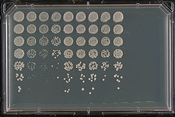
| + | [[File:HTP_pinning_robot.jpg|thumb|right|175px|'''Pinning robot for HTP arrays of yeast''' Credit: ''Wikicommons'']] | ||
| + | By virtue of the high degree of similarity between yeast genes and their human counterparts, and conserved fundamental cellular biology, yeast has become a popular model system for the study of human disease genes. Several approaches have been used to learn more about human genes once a connection between a human and yeast gene is made. In one approach, after a human disease-associated gene is discovered the sequence is compared to the sequences of all genes in the yeast genome to identify the most similar yeast gene(s). To study whether the genes are functionally related, the human gene is then expressed in a yeast stain where the yeast gene has first been inactivated by mutation. This allows researchers to determine whether or not the human gene is able to rescue viability, growth, or more specific defects associated with loss of the yeast gene, a method referred to as functional complementation. If the pathways and/or processes that a yeast gene is involved in are conserved, much can be learned about the function of the human gene based on what is already known about the related yeast gene. Once functional complementation has been established, researchers can use this system to further characterize the function of the related human gene product. Less directed approaches that often utilize high-throughput (HTP) techniques to randomly screen thousands of human genes at one time to identify gene or genes with complementing activity. Such approaches have successfully been used to identify conserved cell cycle regulators (CDC2), genes involved in cancer, and genes involved in neurodegenerative diseases. | ||
| + | |||
| + | [[File:serial_drop_test.jpg|thumb|left|175px|'''Serial drop test with drug of interest''' Credit: ''Wikicommons'']] | ||
| + | |||
| + | |||
| + | There are many scenarios where studies can provide valuable information to researchers about the cellular pathways and/or processes a human gene is involved in when a related yeast gene is not present. For example, some neurodegenerative diseases like Alzheimer’s and Parkinson’s occur as protein aggregates called amyloid accumulate due to protein misfolding and this is toxic to neurons. Studying misfolded yeast proteins with similar amyloid forming potential, called prions, has provided researchers with insight into these neurodegenerative diseases. Alternatively, elevated expression of a disease-associated gene in yeast may result in a phenotype. For example, when expressed at high enough levels, alpha-synuclein, a gene associated with Parkinson’s disease, is toxic. Such a strain can then be used to screen for yeast genes or small molecules that suppress or enhance synuclein-induced toxicity, often providing clues about the relevant cellular pathways. Patients with Amyotrophic Lateral Sclerosis (ALS) or Lou Gehrig’s Disease, often have mutations in a couple of RNA binding proteins which makes them prone to form aggregates that interfere with RNA metabolism. A yeast screen has been used successfully to identify a number of yeast genes with similar properties (form toxic aggregates) providing researchers with new candidate genes to study. Conversely, when expressed in yeast the human RNA binding proteins form toxic aggregates and this strain was used to identify a yeast gene which when mutated blocks the production of these aggregates. | ||
| + | |||
| + | |||
| + | Yeast is becoming the organism of choice in studies aimed at the identification of drug targets and the mode of action of various drugs. Chemogenomics or chemical-genomics refers to the screens that use a combination of chemicals and genomics to probe drug targets and potentially identify novel drugs. Two main approaches have been used in these chemical-genomic studies. In the first, a genome-wide collection of diploid strains is constructed where one of the two identical copies of a gene is deleted, thereby lowering the levels of a particular gene product. Target genes and genes involved in the target pathway become more sensitive to the compound and are preferentially identified in this kind of screen. In a second approach, nonessential genes are systematically deleted and the collection screened with a drug to look for genes which buffer the drug target pathway. This approach is expected to identify genes required for growth in the presence of the compound. Additional approaches using overexpression screens have been used to identify genes involved in drug resistance including the potential drug target. Comparing the expression profile of yeast cells deleted for a gene to those of wild type yeast cells treated with a particular drug can also be an effective way to identify genes which may tell the researchers something about how the drug works in cells. | ||
| + | |||
| + | |||
| + | These are just a few examples of how yeast can be used both aid the study of human disease. Studies in yeast can help researchers learn more about the underlying biology using this model system, or to help them identify drug targets or the drugs mode of action. | ||
| + | |||
| + | ====Resources==== | ||
| + | |||
| + | * [http://www.yeastgenome.org/ ''Saccharomyces'' Genome Database (SGD)] provides comprehensive integrated biological information for the budding yeast ''Saccharomyces cerevisiae'' along with search and analysis tools to explore these data. | ||
| + | |||
| + | * [http://mips.helmholtz-muenchen.de/genre/proj/yeast/ MIPS Comprehensive Yeast Genome Database (CYPD)] presents information on the molecular structure and functional network of the entirely sequenced, well-studied model eukaryote, the budding yeast ''Saccharomyces cerevisiae''. | ||
| + | |||
| + | * [http://www.candidagenome.org/ ''Candida'' Genome Database (CGD)], a resource for genomic sequence data and gene and protein information for ''Candida albicans''. | ||
| + | |||
| + | * [http://www.pombase.org/ PomBase], a comprehensive database for the fission yeast ''Schizosaccharomyces pombe'', providing structural and functional annotation, literature curation and access to large-scale data sets. | ||
| + | |||
| + | * [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3379888/ Fungal genome resources at NCBI], a guide to fungal genome resources at the National Center for Biotechnology Information (NCBI). | ||
| + | |||
| + | * An extensive list of [http://wiki.yeastgenome.org/index.php/External_Links yeast-related resources] on topics ranging from general yeast information to nucleic acids, genomes and proteins, expression data, localization, phenotypes and more. | ||
| + | |||
| + | ====Suggested Reading==== | ||
| + | |||
| + | =====Books===== | ||
| + | |||
| + | * [http://www.genetics.org/site/misc/yeastbook.xhtml YeastBook]. (2011) A comprehensive compendium of reviews that presents the current state of knowledge of the molecular biology, cellular biology, and genetics of the yeast ''Saccharomyces cerevisiae'', Genetics | ||
| + | |||
| + | * [http://books.google.com/books/about/From_a_to_alpha.html?id=79hO0A08odkC From a to alpha: Yeast as a Model for Cellular Differentiation]. (2007) Hiten D. Madhani, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. | ||
| + | |||
| + | * [http://www.cshlpress.com/default.tpl?action=full&cart=133468404249500843&--eqskudatarq=449&newtitle=Landmark%20Papers%20in%20Yeast%20Biology Landmark Papers in Yeast Biology]. (2006), edited by Patrick Linder, David Shore, and Michael N. Hall, Cold Spring Harbor Press, Cold Spring Harbor, New York. | ||
| + | |||
| + | * [http://www.cshlpress.com/default.tpl?action=full&cart=133468404249500843&--eqskudatarq=526&newtitle=Methods%20in%20Yeast%20Genetics%3A%20A%20Cold%20Spring%20%20Harbor%20Laboratory%20Course%20Manual%2C%202005%20Edition Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual]. (2005) David C. Amberg, Daniel J. Burke, and Jeffrey N. Strathern. Cold Spring Laboratory Press, Cold Spring Harbor, New York. | ||
| + | |||
| + | * [http://www.cshlpress.com/default.tpl?cart=133468183249354789&fromlink=T&linkaction=full&linksortby=oop_title&--eqSKUdatarq=676 The Early Days of Yeast Genetics]. (1993) edited by Michael N. Hall and Patrick Linder. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. | ||
| + | |||
| + | * [http://cshmonographs.org/index.php/monographs/issue/view/087969363.21A Volume I: The Molecular and Cellular Biology of the Yeast ''Saccharomyces cerevisiae'': Genome Dynamics, Protein Synthesis, and Energetics]. (1991) edited by James R. Broach, John R. Pringle, and Elizabeth W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. | ||
| + | |||
| + | * [http://cshmonographs.org/index.php/monographs/issue/view/087969365.21B Volume II: The Molecular and Cellular Biology of the Yeast ''Saccharomyces cerevisiae'': Gene Expression]. (1992) edited by Elizabeth W. Jones, John R. Pringle and James R. Broach. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. | ||
| − | [ | + | * [http://cshmonographs.org/index.php/monographs/issue/view/087969364.21C Volume III: The Molecular and Cellular Biology of the Yeast ''Saccharomyces cerevisiae'': Cell Cycle and Cell Biology]. (1997) edited by John R. Pringle, James R. Broach and Elizabeth W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * [http://books.google.com/books/about/Yeast.html?id=-GkHRQAACAAJ Yeast: A Practical Approach]. (1988) edited by I. Campbell and , and John H. Duffus, IRL Press, Ithaca, New York. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | =====Journal Articles===== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | * Duina A.A., Miller M.E., and J.B. Keeney (2014) [http://www.genetics.org/content/197/1/33.long Budding Yeast for Budding Geneticists: A Primer on the ''Saccharomyces cerevisiae'' Model System]. Genetics. '''197''':33-48. | ||
| − | + | * Botstein D. and G.R. Fink (2011) [http://www.genetics.org/content/189/3/695.full.pdf+html Yeast: An Experimental Organism for 21st Century Biology]. Genetics. '''189''':695-704. | |
| − | |||
| − | + | * Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Hitz B.C., Karra K., Krieger C.J., Miyasato S.R., Nash R.S., Park J., Skrzypek M.S., Simison M., Weng S., and E.D. Wong (2011) [http://nar.oxfordjournals.org/content/40/D1/D700.long Saccharomyces Genome Database: the genomics resource of budding yeast]. Nucleic Acids Res. '''40''':D700-D705. | |
| + | <!-- * Cherry J.M., Ball C., Weng S., Juvik, G., Schmidt R., Adler C., Dwight S., Riles L., Mortimer, and D. Botstein (1997) [http://www.ncbi.nlm.nih.gov/pubmed/9169866 Genetic and physical maps of ''Saccharomyces cerevisiae''], Edition 12. Nature '''29''':67-73.--> | ||
| − | = | + | * Altman T.J., Boone C., Churchill G.A., Hengartner M.O., Mackay T.F., and D.L. Stemple (2011) [http://www.ncbi.nlm.nih.gov/pubmed/?term=21765459 The future of model organisms in human disease research]. Nat. Rev. Genet. '''18''':575-582. |
| − | + | * Ho C.H., Piotrowski J., Dixon S.J., Baryshnikova A., Costanzo M., and C. Boone (2011) [http://www.ncbi.nlm.nih.gov/pubmed/?term=21093351 Combining functional genomics and chemical biology to identify targets of bioactive compounds]. Curr Opin Chem Biol. '''15''':66-78. | |
| − | + | * Smith A.M., Ammar R., Nislow C., and G. Giaever (2010) [http://www.ncbi.nlm.nih.gov/pubmed/?term=20546776 A survey of yeast genomic assays for drug and target discovery]. Pharmacol Ther. '''127''':156-164. | |
| − | + | * Dolinski K., and D. Botstein (2007) [http://www.ncbi.nlm.nih.gov/pubmed/?term=17678444 Orthology and functional conservation in eukaryotes]. Annu. Rev. Genet. '''41''':465-507. | |
| − | |||
| − | |||
| − | + | * Barnett J.A. (2007) [http://www.ncbi.nlm.nih.gov/pubmed/?term=17638318 A history of research on yeasts 10: foundations of yeast genetics]. Yeast '''24''':799-845. | |
| − | |||
| − | ''' | ||
| − | + | * Spradling A., Ganetsky B., Hieter P., Johnston M., Olson M., Orr-Weaver T., Rossant J., Sanchez A., and R. Waterston (2006) [http://www.genetics.org/content/172/4/2025.full.pdf+html New roles for model genetic organisms in understanding and treating human disease: report from the 2006 Genetics Society of America meeting]. Genetics '''172''':2025-2032. | |
| − | |||
| − | |||
| − | |||
| − | + | * Sherman F. (2002) [http://www.ncbi.nlm.nih.gov/pubmed/12073320 Getting started with yeast] Methods Enzymol. '''350''':3-41. | |
| − | |||
| − | ( | ||
| − | |||
| − | |||
Latest revision as of 15:04, 16 April 2019
Contents
General Introduction
Most of us know yeast is a very helpful organism, especially with respect to baking, wine making, and brewing. However, what are yeast and why are they the focus of so much research?
Yeast are Fungi
Yeast are single-celled microorganisms that are classified, along with molds and mushrooms, as members of the Kingdom Fungi. Yeasts are evolutionarily diverse and are therefore classified into two separate phyla, Ascomycota or sac fungi and Basidiomycota or higher fungi, that together form the subkingdom Dikarya. Budding yeast, also referred to as “true yeasts”, are members of the phylum Ascomycota and the order Saccharomycetales. Such classifications are based on characteristics of the cell, ascospore, and colony, as well as cellular physiology.
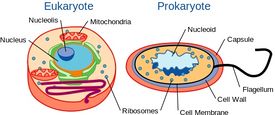
Yeast are Single-celled, but with Cellular Organization Similar to Higher Organisms
Although yeast are single-celled organisms, they possess a cellular organization similar to that of higher organisms, including humans. Specifically, their genetic content is contained within a nucleus. This classifies them as eukaryotic organisms, unlike their single-celled counterparts, bacteria, which do not have a nucleus and are considered prokaryotes.
Natural Habitats
Yeast are widely dispersed in nature with a wide variety of habitats. They are commonly found on plant leaves, flowers, and fruits, as well as in soil. Yeast are also found on the surface of the skin and in the intestinal tracts of warm-blooded animals, where they may live symbiotically or as parasites. The common "yeast infection" is typically caused by Candida albicans. In addition to being the causative agent in vaginal yeast infections, Candida is also the cause of diaper rash and thrush of the mouth and throat.
Why Study Yeast?
Imagine an organism that grows quickly in a flask and whose DNA can be easily manipulated, but also provides insight into basic human biological processes, including disease. Yeast fits that description and is the focus of study for researchers all over the world, resulting in more than 50,000 published scientific articles describing yeast research!
What specific characteristics of yeast make it a “model organism” for study and the focus of so much research? Yeast are single-celled (unicellular) organisms, making them simple to study, but possess a cellular organization similar to that found in higher, multi-cellular organisms such as humans – that is, they possess a nucleus and are therefore eukaryotes, as described above. Most importantly, the similarity in cellular organization between yeast and higher eukaryotes translates to similarities in their fundamental cellular processes so discoveries in yeast frequently provide direct or indirect clues as to how biological processes work in humans.
Another important characteristic of yeast essential to their role as “model organisms” is the fact they are relatively easy to work with. Yeast replicate quickly and are easy to manipulate genetically. The doubling time for yeast (the time required for a cell to duplicate and divide itself) is about 90 minutes. In contrast, human cells growing in culture need about 24 hours to double. There are also well defined genetic methods for yeast that allow researchers to easily isolate mutants, cross them with other mutants or into other genetic backgrounds, and map the locations of genes. In fact, genetic maps constructed based on the genetic distance between genes gave researchers their first view of the genome and its organization, and were the culmination of genetic studies that date back to the first half of the twentieth century.
An accelerated pace of discovery was made possible after the baker's yeast (S. cerevisiae) genome, representing its complete set of genetic material, became the first eukaryotic genome to be sequenced back in 1996. It is smaller and more compact than the human genome (12 million base pairs and ~6,000 genes, compared with 3 billion base pairs and ~20-25,000 protein coding genes). Yet, comparisons of the genomes indicate that ~31% of yeast genes are very similar to human genes and 20% of human disease genes have counterparts in yeast. In addition, yeast cells can exist either as haploids (one set of chromosomes) or diploids (two sets of chromosomes). Since haploids have only one copy of each gene and efficient breaking and rejoining of DNA strands (recombination), it is very easy to delete a specific gene in a haploid and observe the effects on the cell, or the “phenotype” of the deletion mutant. Diploid cells, on the other hand, make it possible to study essential genes (those required for growth and viability) by deleting one copy of the gene and making subtle changes in the other copy. Finally, with the information from the genome sequence an extensive toolkit of molecular reagents and genome-wide collections have been constructed, providing researchers with powerful means to study biological problems. If a yeast gene is known to be similar in DNA sequence to a human gene, studies in yeast can provide powerful clues as to the role of the related gene(s) in humans. Thus, the relative simplicity of studying cellular functions in yeast combined with its relevance to higher organisms makes it a very powerful “model organism” for study.
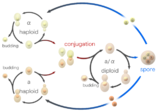
Yeast Life and Cell Cycles
Yeast typically grow asexually by budding. A small bud which will become the daughter cell is formed on the parent (mother) cell, and enlarges with continued grow. As the daughter cell grows, the mother cell duplicates and then segregates its DNA. The nucleus divides and migrates into the daughter cell. Once the bud contains a nucleus and reaches a certain size it separates from the mother cell. The series of events that occur in a cell and lead to duplication and division are referred to as the cell cycle. The cell cycle consists of four distinct phases (G1, S, G2 and M) and is regulated similar to that of the cell cycle in larger eukaryotes. As long as adequate nutrients such as sugar, nitrogen and phosphate are present yeast cells will continue to divide asexually.
Yeast cells can also reproduce sexually. Yeast cells exist as one of two different mating types, a cells and alpha cells. When cells of opposite mating types are mixed together in the lab or randomly come into contact in nature they can mate (conjugate). Before joining the cells change shape in a process called shmooing. The term 'shmoo' was coined based on its similarity in shape to that of a fictional cartoon character of the same name created back in the late 40's by Al Capp, appearing first in his comic strip L'il Abner. During conjugation the shmooing haploid cells first fuse and then their nuclei fuse, resulting in the formation of a diploid cell with two copies of each chromosome. Once formed, diploid cells can reproduce asexually by budding, similar to haploids. However, when diploid cells are starved of nutrients they undergo sporulation. During sporulation diploid cells undergo meiosis, a special form of cell division that reduces the number of chromosomes from two copies back to one copy. After meiosis the haploid nuclei produced in meiosis are packaged into four spores that contain modified cell walls, resulting in structures that are very resistant to environmental stress. These spores can survive long periods of time until conditions become more favorable, such as in the presence of improved nutrients, whereupon they are able to germinate and reproduce asexually. These different states, budding, conjugation and sporulation together make up the yeast life cycle.
Yeast growth and metabolism
When yeast cells are grown in rich carbon sources such as glucose they prefer to grow by fermentation. During fermentation glucose is converted into carbon dioxide and ethanol. Generally, fermentation occurs in the absence of oxygen, and is therefore anaerobic by nature. Even in the presence of oxygen yeast cells prefer to grow fermentatively and this is referred to as the Crabtree Effect after the biologist who discovered this preference. This form of growth is exploited in the making of bread, beer, wine and other alcoholic beverages. Although budding yeast cells prefer to grow by fermentation, when nutrients are limiting they are also able to grow by cellular respiration. During respiration cells convert glucose into carbon dioxide and water, consuming oxygen in the process, and resulting in the production of much larger amounts of energy in the form of ATP.
Historical Discoveries
Yeast has been used as an industrial microorganism for 1000’s of years. The ancient Egyptians used yeast fermentation to leaven bread. There is evidence of grinding stones, baking chambers and drawings of 4000-year-old bakeries. Archaeological digs have uncovered evidence in the form of jars containing the remains of wine that is 7,000 years old.
Yeast were first visualized in 1680 by Antoni van Leeuwenhoek using high quality lenses. However, he thought that these globules were starchy particles of the grain used to make wort, the liquid extract used in brewing, rather than fermenting yeast cells. In 1789, Antoine Lavoisier, a French chemist, contributed to our understanding of the basic chemical reactions needed to produce alcohol from sugarcane. By estimating the proportion of starting materials and products (ethanol and carbon dioxide) after adding yeast paste he concluded that two chemical pathways were used with two thirds of the sugar reduced to alcohol and one third to form carbon dioxide. However, at the time it was thought that yeast were merely there to initiate the reaction rather than being required throughout the process.
In 1815, Joseph-Louis Gay-Lussac, a French chemist, developed methods to maintain grape juice in an unfermented state and discovered that the introduction of ‘ferment’ (which contains yeast) was required to convert unfermented wort, demonstrating the importance of yeast for alcoholic fermentation. In 1835, Charles Cagniard de la Tour used a more powerful microscope to show that yeast were single celled and multiplied by budding. In the 1850s Louis Pasteur discovered that fermented beverages resulted from the conversion of glucose to ethanol by yeast and defined fermentation as "respiration without air". Near the end of the 1800s Eduard Buchner used cell-free extracts obtained by grinding yeast cells to detect zymase, the collection of enzymes that promote or catalyze fermentation and for this he was awarded the Nobel Prize in 1907.
Much of the pioneering work on yeast genetics was carried out by Øjvind Winge. He discovered that yeast alternate between haploid and diploid states and that yeast are heterothallic, as two strains are required to convert haploids to diploids (conjugation). He and his colleague Otto Laustsen devised techniques to micromanipulate yeast so they could be investigated genetically. With this technique, known as "tetrad analysis", a fine needle and a microscope are used to isolate a structure known as an ascus, which contains the four spore products or tetrad resulting from sporulation of a diploid. Once the ascus is isolated, the spores in the tetrad are teased apart and allowed to grow into colonies for genetic analysis. This pioneering work earned him the title ‘The Father of Yeast Genetics’. Some of this work was further clarified by Carl Lindegren, who elucidated the mating-type system in budding yeast, demonstrating the existence of Mat a and Mat alpha cells, devised methods to carry our mass matings between cells of these mating types and used this knowledge to study the genetics of sugar utilization.
Since that time many other researchers have carried out groundbreaking research using budding yeast. Some of these researchers have been awarded the Nobel Prize for significant discoveries made during these studies, including: Dr. Leland Hartwell (2001) for the discovery of genes that regulate the cell cycle (co-winner with Paul Nurse and Tim Hunt); Roger Kornberg (2006) for his studies on the first step of gene expression, the means by which a genes DNA sequence is copied into messenger RNA (mRNA); Drs. Elizabeth Blackburn, Carol Greider and Jack Szostak (2009) for discovering and elucidating the genes and means by which cells protect chromosome ends or telomeres from being degraded; and to Drs. Randy Schekman, James Rothman and Thomas Südhof (2013) for research on the machinery that regulates vesicular traffic. Most recently, Dr. Yoshinori Ohsumi was awarded the prize for his work on autophagy, which began with studies in yeast.
Commercial Applications
Yeast has long been considered to be the organism of choice for the production of alcoholic beverages, bread, and a large variety of industrial products. This is based on the ease with which the metabolism of yeast can be manipulated using genetic techniques, the speed with which it can be grown to high cell yields (biomass), the ease with which this biomass can be separated from products and the knowledge that it is generally recognized as safe (GRAS).
The budding yeast S. cerevisiae and other yeast species have long been used to ferment the sugars of rice, wheat, barley, and corn to produce alcoholic beverages such as beer and wine. There are two major types of brewing yeast, top-fermenting ale yeast and bottom-fermenting lager yeast. Top-fermenting yeast such as S. cerevisiae rise to the surface during fermentation and are used to brew ales, porters, stouts and wheat beers. In contrast, S. pastorianus, (formerly known as S. carlsbergensis) is a bottom-fermenting yeast used to make lager beer. Lager yeasts grow best at lower temperatures. As a result they grow more slowly, produce less surface foam, and therefore typically settle to the bottom of the fermenter. Pilsners, Märzen, Bocks, and American malt liquors are all styles of lager beer. In modern brewing many of the original top fermenting strains have been modified to become bottom fermenters.
Yeast produce wine by fermenting sugars from grape juice (must) into ethanol. Although wine fermentation can be initiated by naturally occurring yeast present in the vineyards, many wineries choose to add a pure yeast culture to dominate and control the fermentation. The bubbles in champagne and sparkling wines are produced by a secondary fermentation, typically in the bottle, which traps the carbon dioxide. Carbon dioxide produced in wine production is released as a by-product. One yeast cell can ferment approximately its own weight in glucose per hour. Under optimal conditions S. cerevisiae can produce up to 18 percent, by volume of ethanol with 15 - 16% being the norm. The sulfur dioxide present in commercially produced wine is added just after the grapes are crushed to kill the naturally present bacteria, mold, and yeast.
Saccharomyces cerevisiae or baker’s yeast has long been used as a leavening agent in baking. Baker’s yeast ferment sugars present in dough, producing carbon dioxide and ethanol. The carbon dioxide becomes trapped in small bubbles in the dough, which causes the dough to rise. Sourdough bread is an exception, as it is not produced using baker's yeast, but is instead made with a combination of wild yeast and bacteria. The yeast Candida milleri is used to strengthen the gluten, and an acid-generating bacteria “Lactobacillus sanfranciscensis”, is used to ferment the maltose.
In addition to these traditional uses yeast has also been used for many other commercial applications. Vegans often use yeast as a cheese substitute and it is often used as a topping for products such as popcorn. It is being used in the petrochemical industry where it has been engineered to produce biofuels such as ethanol, and farnesene, a diesel and jet fuel precursor. It is also used in the production of lubricants and detergents. Yeast is used in the food industry for the production of food additives including colorants, antioxidants, and flavor enhancers. It is the often used in the production of pharmaceuticals including antiparasitics, anticancer compounds, biopharmaceuticals such as insulin, vaccines, and nutraceuticals. Yeast is commonly used in the production of industrial enzymes and chemicals. In the field of environmental bioremediation strains have even been exploited for the removal of metal from mining waste.
Application to Human Disease and Research
By virtue of the high degree of similarity between yeast genes and their human counterparts, and conserved fundamental cellular biology, yeast has become a popular model system for the study of human disease genes. Several approaches have been used to learn more about human genes once a connection between a human and yeast gene is made. In one approach, after a human disease-associated gene is discovered the sequence is compared to the sequences of all genes in the yeast genome to identify the most similar yeast gene(s). To study whether the genes are functionally related, the human gene is then expressed in a yeast stain where the yeast gene has first been inactivated by mutation. This allows researchers to determine whether or not the human gene is able to rescue viability, growth, or more specific defects associated with loss of the yeast gene, a method referred to as functional complementation. If the pathways and/or processes that a yeast gene is involved in are conserved, much can be learned about the function of the human gene based on what is already known about the related yeast gene. Once functional complementation has been established, researchers can use this system to further characterize the function of the related human gene product. Less directed approaches that often utilize high-throughput (HTP) techniques to randomly screen thousands of human genes at one time to identify gene or genes with complementing activity. Such approaches have successfully been used to identify conserved cell cycle regulators (CDC2), genes involved in cancer, and genes involved in neurodegenerative diseases.
There are many scenarios where studies can provide valuable information to researchers about the cellular pathways and/or processes a human gene is involved in when a related yeast gene is not present. For example, some neurodegenerative diseases like Alzheimer’s and Parkinson’s occur as protein aggregates called amyloid accumulate due to protein misfolding and this is toxic to neurons. Studying misfolded yeast proteins with similar amyloid forming potential, called prions, has provided researchers with insight into these neurodegenerative diseases. Alternatively, elevated expression of a disease-associated gene in yeast may result in a phenotype. For example, when expressed at high enough levels, alpha-synuclein, a gene associated with Parkinson’s disease, is toxic. Such a strain can then be used to screen for yeast genes or small molecules that suppress or enhance synuclein-induced toxicity, often providing clues about the relevant cellular pathways. Patients with Amyotrophic Lateral Sclerosis (ALS) or Lou Gehrig’s Disease, often have mutations in a couple of RNA binding proteins which makes them prone to form aggregates that interfere with RNA metabolism. A yeast screen has been used successfully to identify a number of yeast genes with similar properties (form toxic aggregates) providing researchers with new candidate genes to study. Conversely, when expressed in yeast the human RNA binding proteins form toxic aggregates and this strain was used to identify a yeast gene which when mutated blocks the production of these aggregates.
Yeast is becoming the organism of choice in studies aimed at the identification of drug targets and the mode of action of various drugs. Chemogenomics or chemical-genomics refers to the screens that use a combination of chemicals and genomics to probe drug targets and potentially identify novel drugs. Two main approaches have been used in these chemical-genomic studies. In the first, a genome-wide collection of diploid strains is constructed where one of the two identical copies of a gene is deleted, thereby lowering the levels of a particular gene product. Target genes and genes involved in the target pathway become more sensitive to the compound and are preferentially identified in this kind of screen. In a second approach, nonessential genes are systematically deleted and the collection screened with a drug to look for genes which buffer the drug target pathway. This approach is expected to identify genes required for growth in the presence of the compound. Additional approaches using overexpression screens have been used to identify genes involved in drug resistance including the potential drug target. Comparing the expression profile of yeast cells deleted for a gene to those of wild type yeast cells treated with a particular drug can also be an effective way to identify genes which may tell the researchers something about how the drug works in cells.
These are just a few examples of how yeast can be used both aid the study of human disease. Studies in yeast can help researchers learn more about the underlying biology using this model system, or to help them identify drug targets or the drugs mode of action.
Resources
- Saccharomyces Genome Database (SGD) provides comprehensive integrated biological information for the budding yeast Saccharomyces cerevisiae along with search and analysis tools to explore these data.
- MIPS Comprehensive Yeast Genome Database (CYPD) presents information on the molecular structure and functional network of the entirely sequenced, well-studied model eukaryote, the budding yeast Saccharomyces cerevisiae.
- Candida Genome Database (CGD), a resource for genomic sequence data and gene and protein information for Candida albicans.
- PomBase, a comprehensive database for the fission yeast Schizosaccharomyces pombe, providing structural and functional annotation, literature curation and access to large-scale data sets.
- Fungal genome resources at NCBI, a guide to fungal genome resources at the National Center for Biotechnology Information (NCBI).
- An extensive list of yeast-related resources on topics ranging from general yeast information to nucleic acids, genomes and proteins, expression data, localization, phenotypes and more.
Suggested Reading
Books
- YeastBook. (2011) A comprehensive compendium of reviews that presents the current state of knowledge of the molecular biology, cellular biology, and genetics of the yeast Saccharomyces cerevisiae, Genetics
- From a to alpha: Yeast as a Model for Cellular Differentiation. (2007) Hiten D. Madhani, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Landmark Papers in Yeast Biology. (2006), edited by Patrick Linder, David Shore, and Michael N. Hall, Cold Spring Harbor Press, Cold Spring Harbor, New York.
- Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. (2005) David C. Amberg, Daniel J. Burke, and Jeffrey N. Strathern. Cold Spring Laboratory Press, Cold Spring Harbor, New York.
- The Early Days of Yeast Genetics. (1993) edited by Michael N. Hall and Patrick Linder. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Volume I: The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Genome Dynamics, Protein Synthesis, and Energetics. (1991) edited by James R. Broach, John R. Pringle, and Elizabeth W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Volume II: The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Gene Expression. (1992) edited by Elizabeth W. Jones, John R. Pringle and James R. Broach. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Volume III: The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Cell Cycle and Cell Biology. (1997) edited by John R. Pringle, James R. Broach and Elizabeth W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Yeast: A Practical Approach. (1988) edited by I. Campbell and , and John H. Duffus, IRL Press, Ithaca, New York.
Journal Articles
- Duina A.A., Miller M.E., and J.B. Keeney (2014) Budding Yeast for Budding Geneticists: A Primer on the Saccharomyces cerevisiae Model System. Genetics. 197:33-48.
- Botstein D. and G.R. Fink (2011) Yeast: An Experimental Organism for 21st Century Biology. Genetics. 189:695-704.
- Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Hitz B.C., Karra K., Krieger C.J., Miyasato S.R., Nash R.S., Park J., Skrzypek M.S., Simison M., Weng S., and E.D. Wong (2011) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40:D700-D705.
- Altman T.J., Boone C., Churchill G.A., Hengartner M.O., Mackay T.F., and D.L. Stemple (2011) The future of model organisms in human disease research. Nat. Rev. Genet. 18:575-582.
- Ho C.H., Piotrowski J., Dixon S.J., Baryshnikova A., Costanzo M., and C. Boone (2011) Combining functional genomics and chemical biology to identify targets of bioactive compounds. Curr Opin Chem Biol. 15:66-78.
- Smith A.M., Ammar R., Nislow C., and G. Giaever (2010) A survey of yeast genomic assays for drug and target discovery. Pharmacol Ther. 127:156-164.
- Dolinski K., and D. Botstein (2007) Orthology and functional conservation in eukaryotes. Annu. Rev. Genet. 41:465-507.
- Barnett J.A. (2007) A history of research on yeasts 10: foundations of yeast genetics. Yeast 24:799-845.
- Spradling A., Ganetsky B., Hieter P., Johnston M., Olson M., Orr-Weaver T., Rossant J., Sanchez A., and R. Waterston (2006) New roles for model genetic organisms in understanding and treating human disease: report from the 2006 Genetics Society of America meeting. Genetics 172:2025-2032.
- Sherman F. (2002) Getting started with yeast Methods Enzymol. 350:3-41.