Difference between revisions of "SGD Newsletter, April 2018"
(→1011 Genomes at SGD) |
|||
| Line 5: | Line 5: | ||
==1011 Genomes at SGD== | ==1011 Genomes at SGD== | ||
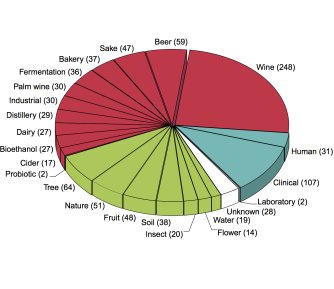
| − | [[File:1011genomes_pie_chart.jpg|thumb|left|upright=1.5| [https://www.nature.com/articles/s41586-018-0030-5 Ecological origins of the 1,011 isolates (from Peter et al., 2018; Creative Commons License]]] | + | [[File:1011genomes_pie_chart.jpg|thumb|left|upright=1.5| [https://www.nature.com/articles/s41586-018-0030-5 Ecological origins of the 1,011 isolates (from Peter et al., 2018; Creative Commons License)]]] |
1,011. That’s the number of different ''Saccharomyces cerevisiae'' yeast strains that were whole-genome sequenced and phenotyped by a team of researchers jointly led by Joseph Schacherer and Gianni Liti and recently published in [https://www.yeastgenome.org/reference/S000216438 Nature]. | 1,011. That’s the number of different ''Saccharomyces cerevisiae'' yeast strains that were whole-genome sequenced and phenotyped by a team of researchers jointly led by Joseph Schacherer and Gianni Liti and recently published in [https://www.yeastgenome.org/reference/S000216438 Nature]. | ||
SGD is happy to announce that in conjunction with the authors and publishers, we are hosting the datasets from the paper at this [https://www.yeastgenome.org/search?q=PMID%3A29643504&category=download&status=Active SGD download site]. These datasets include: the actual genome sequences of the 1,011 isolates; the list of 4,940 common “core” ORFs plus 2,856 ORFs that are variable within the population (together these make up the “pangenome”); copy number variation (CNV) data; phenotyping data for 36 conditions; SNPs and indels relative to the S288C genome; and much more. We hope that the easy availability of these large datasets will be useful to many yeast (and non-yeast) researchers, and as the authors say, will help to “guide future population genomics and genotype–phenotype studies in this classic model system.” | SGD is happy to announce that in conjunction with the authors and publishers, we are hosting the datasets from the paper at this [https://www.yeastgenome.org/search?q=PMID%3A29643504&category=download&status=Active SGD download site]. These datasets include: the actual genome sequences of the 1,011 isolates; the list of 4,940 common “core” ORFs plus 2,856 ORFs that are variable within the population (together these make up the “pangenome”); copy number variation (CNV) data; phenotyping data for 36 conditions; SNPs and indels relative to the S288C genome; and much more. We hope that the easy availability of these large datasets will be useful to many yeast (and non-yeast) researchers, and as the authors say, will help to “guide future population genomics and genotype–phenotype studies in this classic model system.” | ||
| − | <br><br><br><br> | + | <br><br><br><br><br><br><br><br> |
==Yeast Genetics Meeting 2018== | ==Yeast Genetics Meeting 2018== | ||
Revision as of 10:56, 23 April 2018
About this newsletter:
This is the April 2018 issue of the SGD newsletter. The goal of this newsletter is to inform our users about new features in SGD and to foster communication within the yeast community.
Contents
1011 Genomes at SGD
1,011. That’s the number of different Saccharomyces cerevisiae yeast strains that were whole-genome sequenced and phenotyped by a team of researchers jointly led by Joseph Schacherer and Gianni Liti and recently published in Nature.
SGD is happy to announce that in conjunction with the authors and publishers, we are hosting the datasets from the paper at this SGD download site. These datasets include: the actual genome sequences of the 1,011 isolates; the list of 4,940 common “core” ORFs plus 2,856 ORFs that are variable within the population (together these make up the “pangenome”); copy number variation (CNV) data; phenotyping data for 36 conditions; SNPs and indels relative to the S288C genome; and much more. We hope that the easy availability of these large datasets will be useful to many yeast (and non-yeast) researchers, and as the authors say, will help to “guide future population genomics and genotype–phenotype studies in this classic model system.”
Yeast Genetics Meeting 2018
The Yeast Genetics Meeting will be held August 22–26, 2018 at Stanford University in Palo Alto, California. This is the premier meeting for anyone studying eukaryotic biology using the powerful experimental model Saccharomyces cerevisiae #APOYG!. In addition to oral presentations, which are selected from the submitted abstracts, there will also be awards for the best posters. Abstract submission is open now and ends May 17, 2018. This meeting provides a valuable forum for graduate students and postdocs to present their research to an international audience.
The following awards and lectures will be presented at the meeting:
- Lee Hartwell Lecture: Fred Winston, Harvard Medical School
- Ira Herskowitz Award: Mike Cherry, Stanford University
- Lifetime Achievement Award: Steve Oliver, University of Cambridge
- Winge-Lindegren Address: Virginia Zakian, Princeton University
- Special Presentation: David Botstein, Calico
Register now to join us and hundreds of yeast researchers at Stanford University for this exciting four-day meeting. We hope to see you there! #YEAST18
Research Spotlight (Blog posts)
In case you missed them, here are some of the most popular Research Spotlights posted on our blog lately:
- Moonlighting Proteins Do Double Duty Espinosa-Cantú and coworkers investigated which proteins have a catalytic activity and another unrelated job.
- Exploding Yeast to Protect the Wild Maselko and colleagues engineered a yeast strain so that if it doesn't form diploids within the same strain the diploids will explode!
- Strung Out Chromosomes and Coyotes Buehl and colleagues provide evidence that if H3 is not acetylated by Gcn5p, the H3 tail can act as a secondary anchor if the tension sensing motif is mutated.
- Don't Ignore the Scullery Maid or Pol III Filer and coworkers show that Pol III plays a critical role in longevity, and like Cinderella, should not be ignored!
- Trouble with Triplets Koch and coworkers found that ISW1 is important for making sure that nucleosomes get placed back into DNA after they are removed to make room for Pol II.
SGD YouTube Channel
SGD is actively expanding our library of short video tutorials to help you use various SGD tools and pages. We've also started making videos that highlight exciting new research in the yeast community. Check out our latest Research Spotlight videos:
- Trouble with Triplets
- Now Yeast Even Finds Fungal Pathogens!
- Exploding Yeast to Protect the Wild
- A SRTain Surprise in a Lipid Droplet
Videos are accessible via SGD's YouTube channel. If you have a great idea for a video, or would like to see a particular topic covered, feel free to contact us!
Recent Publications from SGD Staff
- Engel SR, Skrzypek M, Hellerstedt ST, Wong ED, Nash RS, Weng S, Binkley G, Sheppard TK, Karra K, Cherry JM (2018). Updated regulation curation model at the Saccharomyces Genome Database. Database (Oxford). January 2018, bay007.
What else have we been up to lately?
You may have seen SGD representatives at the following meetings:
- PI Mike Cherry, senior biocuration scientists Stacia Engel and Edith Wong, and associate biocuration scientist Patrick Ng attended and presented talks and posters at The 11th International Biocuration Conference in Shanghai, China in April 2018.
Upcoming Meetings
EMBO Workshop Gene transcription in yeast: From global analyses to single cells
Sant Feliu de Guixols, Spain
June 9 - 14, 2018
Registration deadline: March 12, 2018
RCN-UBE: Yeast ORFan Gene Project: Finding a Place for ORFans to GO (Summer Workshop)
Rhodes College, Memphis TN
June 11 - 15, 2018
Application submission deadline: February 26, 2018
Yeast Chromosome Biology and Cell Cycle
Steamboat Grand, Steamboat Springs, CO
July 15 - 20, 2018
Application submission deadline: June 13, 2018
Yeast Genetics & Genomics Course
Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
July 24 - August 13, 2018
Application Deadline: April 15, 2018
5th International Synthetic & Systems Biology Summer School - SSBSS 2018
Certosa di Pontignano (Siena) Tuscany, Italy
July 25 - 29, 2018
Application and abstract submission deadline: March 31, 2018
Yeast Genetics Meeting
Stanford University, Palo Alto, California
August 22 – 26, 2018
13th International Meeting on Yeast Ageing and Apoptosis
The Irish College, Leuven, Belgium
August 26 – 30, 2018
13th Edition of the International Levures, Modèles et Outils (Yeasts, Models and Tools)
Musikinsel, Rheinau, Switzerland
September 11-14, 2018
International Specialized Symposium on Yeasts, "Yeast Odyssey: from nature to industry"
Bariloche, Patagonia, Argentina
October 1 - 4, 2018
International Workshop on Brewing Yeasts
Bariloche, Patagonia, Argentina
October 5 - 6, 2018
EMBO Workshop on Experimental Approaches to Evolution and Ecology Using Yeast and Other Model Systems
Heidelberg, Germany
October 17 - 20, 2018